+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4abb | ||||||
|---|---|---|---|---|---|---|---|
| Title | Fragments bound to bovine trypsin for the SAMPL challenge | ||||||
 Components Components | CATIONIC TRYPSIN | ||||||
 Keywords Keywords | HYDROLASE / FRAGMENT SCREENING / MODELLING | ||||||
| Function / homology |  Function and homology information Function and homology informationtrypsin / serpin family protein binding / serine protease inhibitor complex / digestion / endopeptidase activity / serine-type endopeptidase activity / proteolysis / extracellular space / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.25 Å MOLECULAR REPLACEMENT / Resolution: 1.25 Å | ||||||
 Authors Authors | Newman, J. / Peat, T.S. | ||||||
 Citation Citation |  Journal: J.Comput.Aided Mol.Des. / Year: 2012 Journal: J.Comput.Aided Mol.Des. / Year: 2012Title: The Dingo Dataset: A Comprehensive Set of Data for the Sampl Challenge. Authors: Newman, J. / Dolezal, O. / Fazio, V. / Caradoc-Davies, T. / Peat, T.S. #1: Journal: J.Biomol.Screen / Year: 2009 Title: Practical Aspects of the Sampl Challenge: Providing an Extensive Experimental Data Set for the Modeling Community. Authors: Newman, J. / Fazio, V.J. / Caradoc-Davies, T.T. / Branson, K. / Peat, T.S. #2:  Journal: Aust.J.Chem. / Year: 2013 Journal: Aust.J.Chem. / Year: 2013Title: Fragment Screening for the Modelling Community: Spr, Itc, and Crystallography Authors: Dolezal, O. / Doughty, L. / Hattarki, M.K. / Fazio, V.J. / Caradoc-Davies, T.T. / Newman, J. / Peat, T.S. | ||||||
| History |
| ||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AB" IN EACH CHAIN ON SHEET RECORDS BELOW ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AB" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4abb.cif.gz 4abb.cif.gz | 78.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4abb.ent.gz pdb4abb.ent.gz | 59.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4abb.json.gz 4abb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4abb_validation.pdf.gz 4abb_validation.pdf.gz | 472.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4abb_full_validation.pdf.gz 4abb_full_validation.pdf.gz | 479.9 KB | Display | |
| Data in XML |  4abb_validation.xml.gz 4abb_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  4abb_validation.cif.gz 4abb_validation.cif.gz | 25.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ab/4abb https://data.pdbj.org/pub/pdb/validation_reports/ab/4abb ftp://data.pdbj.org/pub/pdb/validation_reports/ab/4abb ftp://data.pdbj.org/pub/pdb/validation_reports/ab/4abb | HTTPS FTP |
-Related structure data
| Related structure data |  4ab8C  4ab9C  4abaC  4abdC  4abeC  4abfC  4abgC  4abhC  1k1mS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 23324.287 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: SIGMA / Source: (natural)  |
|---|
-Non-polymers , 6 types, 356 molecules 



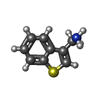






| #2: Chemical | ChemComp-SO4 / | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-CA / | ||||||
| #4: Chemical | | #5: Chemical | ChemComp-DMS / | #6: Chemical | ChemComp-K9S / | #7: Water | ChemComp-HOH / | |
-Details
| Has protein modification | Y |
|---|---|
| Nonpolymer details | LIGAND K9S FRAGMENT CC12313 FROM MAYBRIDGE |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1000 X-RAY DIFFRACTION / Number of used crystals: 1000 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.92 Å3/Da / Density % sol: 36.1 % / Description: NONE |
|---|---|
| Crystal grow | pH: 5.8 Details: 22.5% PEG 3350, 0.18 M AMMONIUM SULFATE, 0.12 M SODIUM THIOCYANATE, 0.09 M BIS-TRIS PH 5.5, 0.01 M TRIS PH 8.5 (FINAL MEASURED PH=5.82). THE PROTEIN WAS AT 2 MM (47 MG/ML), WITH 4 MM ...Details: 22.5% PEG 3350, 0.18 M AMMONIUM SULFATE, 0.12 M SODIUM THIOCYANATE, 0.09 M BIS-TRIS PH 5.5, 0.01 M TRIS PH 8.5 (FINAL MEASURED PH=5.82). THE PROTEIN WAS AT 2 MM (47 MG/ML), WITH 4 MM BENZYLAMINE AND 10 MM CALCIUM CHLORIDE ADDED TO STABILIZE IT. THE CRYSTALLIZATIONS WERE SET UP WITH A PHOENITO PROTOCOL (NEWMAN ET AL. 2008), WHERE A PHOENIX ROBOT (ART ROBBINS INSTRUMENTS, SUNNYSIDE, CA) WAS USED TO DISPENSE THE PROTEIN INTO AN SD2 CRYSTALLIZATION PLATE (PRE-FILLED WITH 50 ML RESERVOIR SOLUTION) AND A MOSQUITO ROBOT (TTP LABTECH, MELBOURN, UK) WAS USED TO DISPENSE THE RESERVOIR SOLUTION AND SEED STOCK OVER THE PROTEIN DROPLET. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX1 / Wavelength: 0.95367 / Beamline: MX1 / Wavelength: 0.95367 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Aug 23, 2009 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.95367 Å / Relative weight: 1 |
| Reflection | Resolution: 1.25→43.9 Å / Num. obs: 59467 / % possible obs: 99.9 % / Observed criterion σ(I): 1 / Redundancy: 7 % / Rmerge(I) obs: 0.058 / Net I/σ(I): 21.6 |
| Reflection shell | Resolution: 1.25→1.32 Å / Redundancy: 6.2 % / Rmerge(I) obs: 0.24 / Mean I/σ(I) obs: 7.4 / % possible all: 99.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1K1M Resolution: 1.25→43.91 Å / Cor.coef. Fo:Fc: 0.97 / Cor.coef. Fo:Fc free: 0.964 / SU B: 0.488 / SU ML: 0.022 / Cross valid method: THROUGHOUT / ESU R: 0.04 / ESU R Free: 0.04 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.U VALUES REFINED INDIVIDUALLY.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 8.738 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.25→43.91 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller








































































































































































































































































 PDBj
PDBj





