[English] 日本語
 Yorodumi
Yorodumi- PDB-1sfi: High resolution structure of a potent, cyclic protease inhibitor ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1sfi | ||||||
|---|---|---|---|---|---|---|---|
| Title | High resolution structure of a potent, cyclic protease inhibitor from sunflower seeds | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / TRYPSIN INHIBITOR / CYCLIC PEPTIDE / PROTEASE / BOVINE-TRYPSIN / SERINE PROTEASE-INHIBITOR / HYDROLASE-HYDROLASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of endopeptidase activity / endopeptidase inhibitor activity / trypsin / serpin family protein binding / serine protease inhibitor complex / digestion / serine-type endopeptidase inhibitor activity / protease binding / endopeptidase activity / serine-type endopeptidase activity ...negative regulation of endopeptidase activity / endopeptidase inhibitor activity / trypsin / serpin family protein binding / serine protease inhibitor complex / digestion / serine-type endopeptidase inhibitor activity / protease binding / endopeptidase activity / serine-type endopeptidase activity / proteolysis / extracellular space / metal ion binding Similarity search - Function | ||||||
| Biological species |   | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.65 Å MOLECULAR REPLACEMENT / Resolution: 1.65 Å | ||||||
 Authors Authors | Luckett, S. / Garcia, R.S. / Barker, J.J. / Konarev, A.V. / Shewry, P. / Clarke, A.R. / Brady, R.L. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. Authors: Luckett, S. / Garcia, R.S. / Barker, J.J. / Konarev, A.V. / Shewry, P.R. / Clarke, A.R. / Brady, R.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1sfi.cif.gz 1sfi.cif.gz | 67 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1sfi.ent.gz pdb1sfi.ent.gz | 47.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1sfi.json.gz 1sfi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sf/1sfi https://data.pdbj.org/pub/pdb/validation_reports/sf/1sfi ftp://data.pdbj.org/pub/pdb/validation_reports/sf/1sfi ftp://data.pdbj.org/pub/pdb/validation_reports/sf/1sfi | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1tldS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 23324.287 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #2: Protein/peptide | 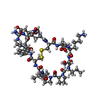  Type: Polypeptide / Class: Trypsin inhibitor / Mass: 1535.829 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: CYCLIC PEPTIDE / Source: (natural) Type: Polypeptide / Class: Trypsin inhibitor / Mass: 1535.829 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Details: CYCLIC PEPTIDE / Source: (natural)  | ||||||||
| #3: Chemical | | #4: Chemical | ChemComp-CA / | #5: Water | ChemComp-HOH / | Compound details | CYCLIC PEPTIDE BOUND TO ACTIVE SITE OF TRYPSIN NO 3'-TERMINAL RESIDUE, CYCLIC PEPTIDE | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.75 Å3/Da / Density % sol: 54.8 % | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 8 / Details: pH 8.0 | |||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion | |||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SRS SRS  / Beamline: PX7.2 / Wavelength: 1.448 / Beamline: PX7.2 / Wavelength: 1.448 |
| Detector | Detector: IMAGE PLATE / Date: May 1, 1998 / Details: MIRRORS |
| Radiation | Monochromator: SI(111) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.448 Å / Relative weight: 1 |
| Reflection | Resolution: 1.65→20 Å / Num. obs: 32839 / % possible obs: 96.7 % / Redundancy: 2.5 % / Biso Wilson estimate: 18.43 Å2 / Rmerge(I) obs: 0.067 / Net I/σ(I): 14 |
| Reflection shell | Resolution: 1.65→1.78 Å / Redundancy: 2.2 % / Rmerge(I) obs: 0.223 / Mean I/σ(I) obs: 3.8 / % possible all: 91 |
| Reflection | *PLUS Num. measured all: 82087 |
| Reflection shell | *PLUS % possible obs: 91 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1TLD Resolution: 1.65→20 Å / SU B: 1.67 / SU ML: 0.06 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.09 / ESU R Free: 0.09
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.11 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.65→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor Rfree: 0.203 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller





















 PDBj
PDBj






