[English] 日本語
 Yorodumi
Yorodumi- PDB-1gi3: A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTER... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1gi3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | ||||||
 Components Components | BETA-TRYPSIN | ||||||
 Keywords Keywords | HYDROLASE / three-centered / very short hydrogen bond / oxyanion hole water / shift of pKa of His57 / structure-based drug design / specificity / urokinase / trypsin / thrombin / Zn+2-mediated inhibition | ||||||
| Function / homology |  Function and homology information Function and homology informationtrypsin / serpin family protein binding / serine protease inhibitor complex / digestion / endopeptidase activity / serine-type endopeptidase activity / proteolysis / extracellular space / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  FOURIER SYNTHESIS / Resolution: 1.44 Å FOURIER SYNTHESIS / Resolution: 1.44 Å | ||||||
 Authors Authors | Katz, B.A. / Elrod, K. / Luong, C. / Rice, M. / Mackman, R.L. / Sprengeler, P.A. / Spencer, J. / Hatayte, J. / Janc, J. / Link, J. ...Katz, B.A. / Elrod, K. / Luong, C. / Rice, M. / Mackman, R.L. / Sprengeler, P.A. / Spencer, J. / Hatayte, J. / Janc, J. / Link, J. / Litvak, J. / Rai, R. / Rice, K. / Sideris, S. / Verner, E. / Young, W. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2001 Journal: J.Mol.Biol. / Year: 2001Title: A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site. Authors: Katz, B.A. / Elrod, K. / Luong, C. / Rice, M.J. / Mackman, R.L. / Sprengeler, P.A. / Spencer, J. / Hataye, J. / Janc, J. / Link, J. / Litvak, J. / Rai, R. / Rice, K. / Sideris, S. / Verner, E. / Young, W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1gi3.cif.gz 1gi3.cif.gz | 111.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1gi3.ent.gz pdb1gi3.ent.gz | 87.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1gi3.json.gz 1gi3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gi/1gi3 https://data.pdbj.org/pub/pdb/validation_reports/gi/1gi3 ftp://data.pdbj.org/pub/pdb/validation_reports/gi/1gi3 ftp://data.pdbj.org/pub/pdb/validation_reports/gi/1gi3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1ghvC  1ghwC  1ghxC  1ghyC  1ghzC  1gi0C  1gi1C 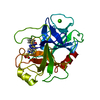 1gi2C  1gi4C  1gi5C  1gi6C  1gi7C  1gi8C  1gi9C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 23324.287 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #2: Chemical | ChemComp-CA / |
| #3: Chemical | ChemComp-BMZ / |
| #4: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.99 Å3/Da / Density % sol: 58.83 % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion / pH: 8.1 Details: magnesium sulfate soak at pH 9.52. vapor diffusion at 298 K, pH 8.10 | |||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / PH range low: 8.2 / PH range high: 7.5 | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 298 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: May 18, 2000 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.32→47.22 Å / Num. all: 65870 / Num. obs: 40323 / % possible obs: 61.2 % / Observed criterion σ(I): 0.8 / Redundancy: 3 % / Rmerge(I) obs: 0.095 / Net I/σ(I): 7.4 |
| Reflection shell | Resolution: 1.44→1.51 Å / Rmerge(I) obs: 0.255 / Num. unique all: 2334 / % possible all: 37.2 |
| Reflection | *PLUS Num. measured all: 110074 |
- Processing
Processing
| Software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS / Resolution: 1.44→7.5 Å / σ(F): 1.8 / Stereochemistry target values: X-PLOR force field FOURIER SYNTHESIS / Resolution: 1.44→7.5 Å / σ(F): 1.8 / Stereochemistry target values: X-PLOR force fieldDetails: Residues simultaneously refined in two or more conformations are: Val53, Leu66, Ser86, Ser113, Ser130, Lys159, Asp165, Ser170, Ser217, Lys230, Ser236 Note that HOH383 makes short H-bonds to ...Details: Residues simultaneously refined in two or more conformations are: Val53, Leu66, Ser86, Ser113, Ser130, Lys159, Asp165, Ser170, Ser217, Lys230, Ser236 Note that HOH383 makes short H-bonds to OgSer195 and O6' of the inhibitor Disordered waters are: HOH304 which is close to HOH305; HOH331 which is close to HOH332; HOH398 which is close to HOH399; HOH407 which is close to HOH408 which is close to HOH409; HOH468 which is close to HOH469 which is close to HOH470; HOH537 which is close to HOH538; HOH618 which is close to HOH619; HOH719 which is close to HOH720 which is close to HOH721; HOH838 which is close to HOH839; HOH917 which is close to HOH918; His40 and HIS91 are MONOPROTONATED ON THE EPSILON NITROGEN. His57 is doubly protonated.
| ||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.44→7.5 Å
| ||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 7.5 Å / σ(F): 1.8 / % reflection Rfree: 10 % / Rfactor all: 0.206 / Rfactor obs: 0.202 | ||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 1.44 Å / Lowest resolution: 1.51 Å / Rfactor Rfree: 0.235 / Rfactor obs: 0.206 |
 Movie
Movie Controller
Controller























 PDBj
PDBj