4OOM
 
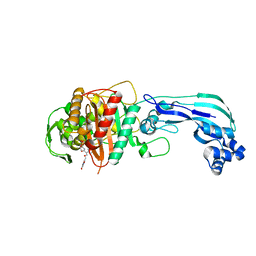 | | Crystal structure of PBP3 in complex with BAL30072 ((2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide) | | Descriptor: | (2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
4PY1
 
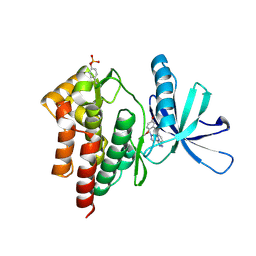 | | Crystal structure of Tyk2 in complex with compound 15, 6-((2,5-dimethoxyphenyl)thio)-3-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine | | Descriptor: | 6-[(2,5-dimethoxyphenyl)sulfanyl]-3-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazine, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Kinase domain inhibition of leucine rich repeat kinase 2 (LRRK2) using a [1,2,4]triazolo[4,3-b]pyridazine scaffold.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6MNY
 
 | |
3TUB
 
 | |
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation
To Be Published
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation
To Be Published
|
|
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
8E50
 
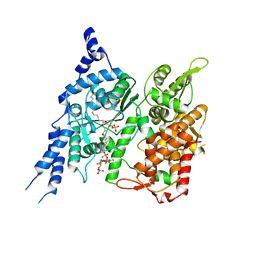 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, COENZYME A, Glycerol-3-phosphate acyltransferase 1, ... | | Authors: | Wasilko, D.J, Johnson, Z.L, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8E4Y
 
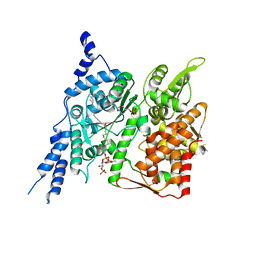 | | Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase 1, mitochondrial, ... | | Authors: | Johnson, Z.L, Wasilko, D.J, Ammirati, M, Chang, J.S, Han, S, Wu, H. | | Deposit date: | 2022-08-19 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the acyl-transfer mechanism of human GPAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8TDW
 
 | | ssRNA bound SAMHD1 T open | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, RNA (5'-R(P*CP*CP*GP*AP*CP*C)-3'), ... | | Authors: | Sung, M, Huynh, K, Han, S. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Guanine-containing ssDNA and RNA induce dimeric and tetrameric structural forms of SAMHD1.
Nucleic Acids Res., 51, 2023
|
|
8TDV
 
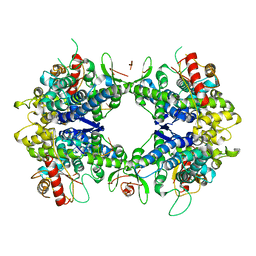 | | ssRNA bound SAMHD1 T closed | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, RNA (5'-R(P*CP*CP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Sung, M, Huynh, K, Han, S. | | Deposit date: | 2023-07-05 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Guanine-containing ssDNA and RNA induce dimeric and tetrameric structural forms of SAMHD1.
Nucleic Acids Res., 51, 2023
|
|
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
7UJA
 
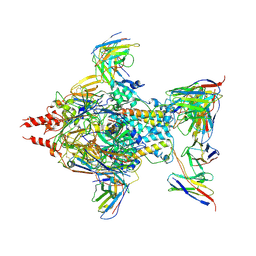 | | Cryo-EM structure of Human respiratory syncytial virus F variant (construct pXCS847A) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM14 Fab heavy chain, AM14 Fab light chain, ... | | Authors: | Lees, J.A, Ammirati, M, Han, S. | | Deposit date: | 2022-03-30 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rational design of a highly immunogenic prefusion-stabilized F glycoprotein antigen for a respiratory syncytial virus vaccine.
Sci Transl Med, 15, 2023
|
|
4E3R
 
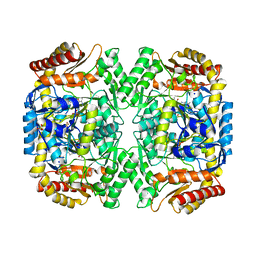 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4E3Q
 
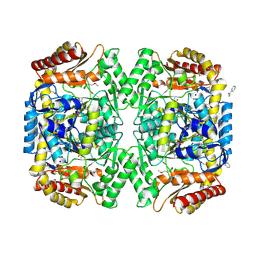 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
6USF
 
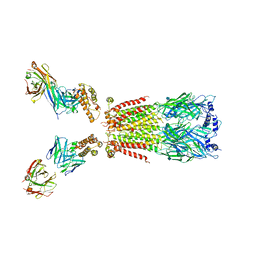 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
6UR8
 
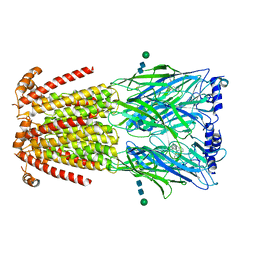 | | CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chimera of soluble cytochrome b562 (BRIL) and neuronal acetylcholine receptor subunit alpha-4, Neuronal acetylcholine receptor subunit beta-2, ... | | Authors: | Alvarez, F.J.D, Mukherjee, S, Han, S, Ammirati, M, Kossiakoff, A.A. | | Deposit date: | 2019-10-22 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
8TB0
 
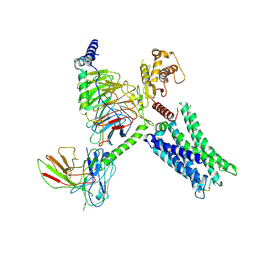 | | Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 | | Descriptor: | GPR61 fused to dominant negative G alpha S/I N18 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8TB7
 
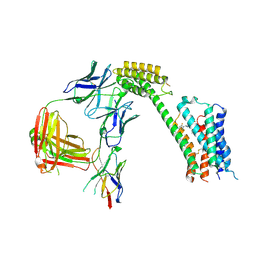 | | Cryo-EM Structure of GPR61- | | Descriptor: | 6-{[(3,5-difluoropyridin-4-yl)methyl]amino}-N-(4-ethoxy-6-methylpyrimidin-2-yl)-2-methoxy-N-(2-methoxyethyl)pyridine-3-sulfonamide, Fab hinge-binding nanobody, Fab24 BAK5 heavy chain, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
6NZU
 
 | | Structure of the human frataxin-bound iron-sulfur cluster assembly complex | | Descriptor: | Acyl carrier protein, Cysteine desulfurase, mitochondrial, ... | | Authors: | Fox, N.G, Yu, X, Xidong, F, Alain, M, Joseph, N, Claire, S.D, Christine, B, Han, S, Yue, W.W. | | Deposit date: | 2019-02-14 | | Release date: | 2019-05-22 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism.
Nat Commun, 10, 2019
|
|
4ZQS
 
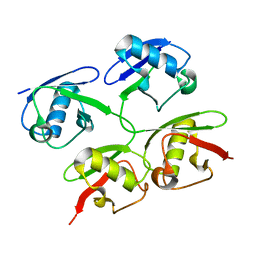 | | New compact conformation of linear Ub2 structure | | Descriptor: | ubiquitin | | Authors: | Thach, T.T, Shin, D, Han, S, Lee, S. | | Deposit date: | 2015-05-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | New conformations of linear polyubiquitin chains from crystallographic and solution-scattering studies expand the conformational space of polyubiquitin.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7UJN
 
 | | Structure of Human SAMHD1 with Non-Hydrolysable dGTP Analog | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | | Authors: | Huynh, K.W, Ammirati, M, Han, S. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Phosphorylation of SAMHD1 Thr592 increases C-terminal domain dynamics, tetramer dissociation and ssDNA binding kinetics.
Nucleic Acids Res., 50, 2022
|
|
6B5V
 
 | | Structure of TRPV5 in complex with econazole | | Descriptor: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Lodowski, D.T, Huynh, K.W, Yazici, A, del Rosario, J, Kapoor, A, Basak, S, Samanta, A, Chakrapani, S, Zhou, Z.H, Filizola, M, Rohacs, T, Han, S, Moiseenkova-Bell, V.Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7L7K
 
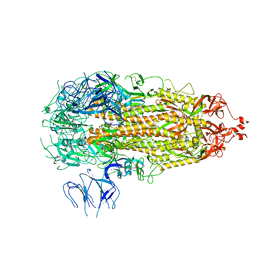 | |
7L7F
 
 | |
