5KHX
 
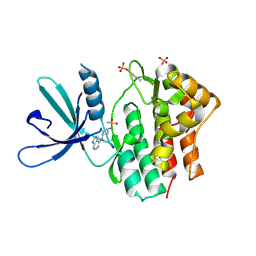 | | Crystal structure of JAK1 in complex with PF-4950736 | | Descriptor: | Tyrosine-protein kinase JAK1, ~{N}-[3-[methyl(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl]methanesulfonamide | | Authors: | Han, S, Caspers, N.L. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of a high-throughput crystal structure-determination platform for JAK1 using a novel metal-chelator soaking system.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
4DD8
 
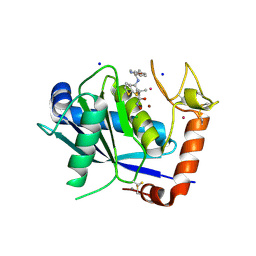 | | ADAM-8 metalloproteinase domain with bound batimastat | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hall, T, Shieh, H.S, Day, J.E, Caspers, N, Chrencik, J.E, Williams, J.M, Pegg, L.E, Pauley, A.M, Moon, A.F, Krahn, J.M, Fischer, D.H, Kiefer, J.R, Tomasselli, A.G, Zack, M.D. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ADAM-8 catalytic domain complexed with batimastat.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3LJT
 
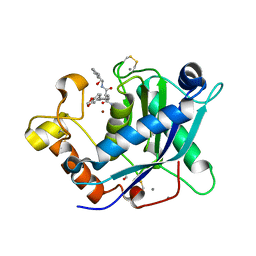 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (2R)-2-[4-(1,3-benzodioxol-5-yl)benzyl]-N~4~-hydroxy-N~1~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]butanediamide, 1,2-ETHANEDIOL, A disintegrin and metalloproteinase with thrombospondin motifs 5, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N. | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure analysis reveals the flexibility of the ADAMTS-5 active site.
Protein Sci., 20, 2011
|
|
5KHW
 
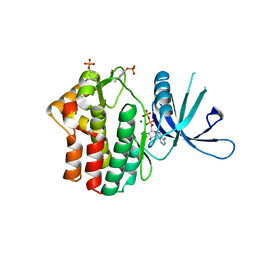 | | Crystal structure of JAK1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Tyrosine-protein kinase JAK1 | | Authors: | Han, S, Caspers, N.L. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Development of a high-throughput crystal structure-determination platform for JAK1 using a novel metal-chelator soaking system.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6MNY
 
 | |
4HCU
 
 | | Crystal structure of ITK in complext with compound 40 | | Descriptor: | 3-{4-amino-1-[(3R)-1-propanoylpiperidin-3-yl]-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-[4-(propan-2-yl)phenyl]benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
4HCT
 
 | | Crystal structure of ITK in complex with compound 52 | | Descriptor: | 3-{1-[(3R)-1-acryloylpiperidin-3-yl]-4-amino-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-(3-tert-butylphenyl)benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Zapf, C.W, Gerstenberger, B.S, Xing, L, Limburg, D.C, Anderson, D.R, Caspers, N, Han, S, Aulabaugh, A, Kurumbail, R, Shakya, S, Li, X, Spaulding, V, Czerwinski, R.M, Seth, N, Medley, Q.G. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
4HCV
 
 | | Crystal structure of ITK in complex with compound 53 | | Descriptor: | 3-{4-amino-1-[(3S)-1-propanoylpiperidin-3-yl]-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-[4-(propan-2-yl)phenyl]benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
3HY9
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (3R)-N~2~-(cyclopropylmethyl)-N~1~-hydroxy-3-(3-hydroxybenzyl)-N~4~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-L-aspartamide, CALCIUM ION, Catalytic Domain of ADAMTS-5, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3FZ1
 
 | | Crystal structure of a benzthiophene inhibitor bound to human Cyclin-dependent Kinase-2 (CDK-2) | | Descriptor: | (3R)-3-(aminomethyl)-9-methoxy-1,2,3,4-tetrahydro-5H-[1]benzothieno[3,2-e][1,4]diazepin-5-one, Cell division protein kinase 2 | | Authors: | Kurumbail, R.G, Caspers, N. | | Deposit date: | 2009-01-23 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Benzothiophene inhibitors of MK2. Part 2: improvements in kinase selectivity and cell potency.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HYG
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (2R)-N~4~-hydroxy-2-(3-hydroxybenzyl)-N~1~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3HY7
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with Marimastat | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3FYK
 
 | | Crystal structure of a benzthiophene lead bound to MAPKAP Kinase-2 (MK-2) | | Descriptor: | (3R)-3-(aminomethyl)-9-methoxy-1,2,3,4-tetrahydro-5H-[1]benzothieno[3,2-e][1,4]diazepin-5-one, MAP kinase-activated protein kinase 2 | | Authors: | Kurumbail, R.G, Caspers, N. | | Deposit date: | 2009-01-22 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Benzothiophene inhibitors of MK2. Part 2: improvements in kinase selectivity and cell potency.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FYJ
 
 | | Crystal structure of an optimzied benzothiophene inhibitor bound to MAPKAP Kinase-2 (MK-2) | | Descriptor: | (10R)-10-methyl-3-(6-methylpyridin-3-yl)-9,10,11,12-tetrahydro-8H-[1,4]diazepino[5',6':4,5]thieno[3,2-f]quinolin-8-one, MAP kinase-activated protein kinase 2 | | Authors: | Kurumbail, R.G, Caspers, N. | | Deposit date: | 2009-01-22 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Benzothiophene inhibitors of MK2. Part 2: improvements in kinase selectivity and cell potency.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3T9T
 
 | | Crystal structure of BTK mutant (F435T,K596R) complexed with Imidazo[1,5-a]quinoxaline | | Descriptor: | (2Z)-4-(dimethylamino)-N-{7-fluoro-4-[(2-methylphenyl)amino]imidazo[1,5-a]quinoxalin-8-yl}-N-methylbut-2-enamide, GLYCEROL, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imidazo[1,5-a]quinoxalines as irreversible BTK inhibitors for the treatment of rheumatoid arthritis.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2P3G
 
 | | Crystal structure of a pyrrolopyridine inhibitor bound to MAPKAP Kinase-2 | | Descriptor: | 2-[2-(2-FLUOROPHENYL)PYRIDIN-4-YL]-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 2 | | Authors: | Kurumbail, R.G, Caspers, N. | | Deposit date: | 2007-03-08 | | Release date: | 2007-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Pyrrolopyridine Inhibitors of Mitogen-Activated Protein Kinase-Activated Protein Kinase 2 (MK-2).
J.Med.Chem., 50, 2007
|
|
4M14
 
 | |
4M15
 
 | |
4M0Y
 
 | |
4M13
 
 | |
4M12
 
 | |
4M0Z
 
 | |
4OOM
 
 | | Crystal structure of PBP3 in complex with BAL30072 ((2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide) | | Descriptor: | (2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
4OON
 
 | | Crystal structure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) | | Descriptor: | (4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid, Penicillin-binding protein 1A | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
4OOL
 
 | | Crystal structure of PBP3 in complex with compound 14 ((2E)-2-({[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}acetyl]amino}-3-oxopropyl]oxy}imino)pentanedioic acid) | | Descriptor: | (2E)-2-({[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}acetyl]amino}-3-oxopropyl]oxy}imino)pentanedioic acid, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
