Derived databases
eF-site
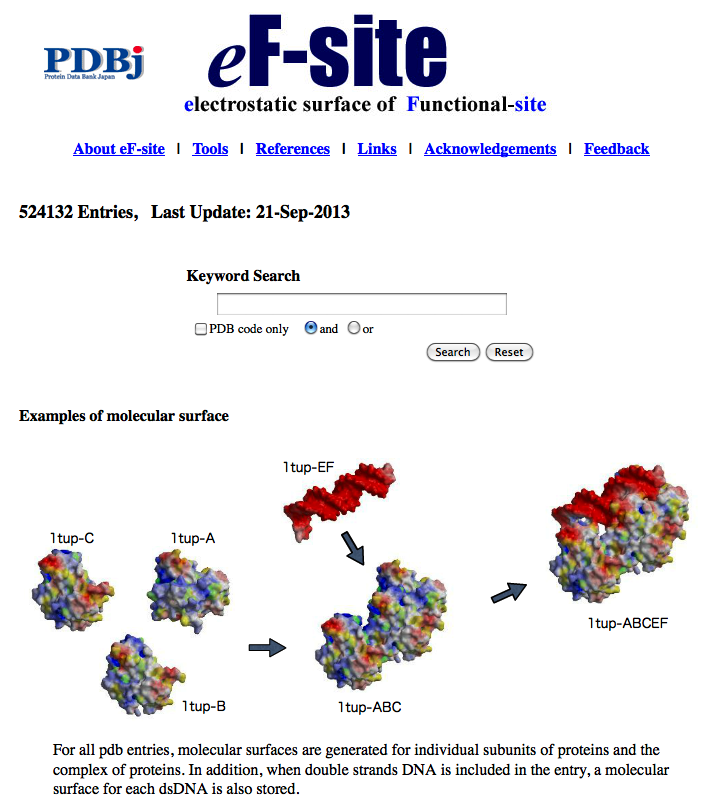
eF-site (electrostatic-surface of Functional site) is a database for molecular surfaces of proteins' functional sites, displaying the electrostatic potentials and hydrophobic properties together on the Connolly surfaces of the active sites, for analyses of the molecular recognition mechanisms.
eF-surf

eF-surf is a web server to calculate the molecular surface of the up-loaded file with PDB format in the same way as the eF-site database.
eF-seek

eF-seek is a web server to search for the similar ligand binding sites for the uploaded coordinate file with PDB format. The representative binding sites in eF-site database are search by our own algorithm based on the clique search algorithm.
ProMode Elastic
ProMode Elasticis a database of normal mode analysis of PDB data. The normal mode analysis is performed by the program PDBETA we have developed. PDBETA is a program of Elastic-network-model based normal mode analysis in Torsional Angle space for PDB data. PDBETA can describe molecular structures with relatively smaller number of degrees of freedom, and take into computation not only proteins but also DNA, RNA, and ligand molecules (hydrogen atoms and water molecules are excluded currently to suppress the number of variables).







