

[Introduction | Basic Usage | Manual | Graphics Examples | Download & Install | Acknowledgment | jV wiki | PDBj Home ]
You can execute basic operations such as rotation and translation with mouse action. Major actions are described in the following table:
| Action | Mouse Operation | |
|---|---|---|
| Windows, Linux | Mac OS X | |
| rotate around X (horizontal) axis rotate around Y (vertical) axis |
left drag | drag |
| rotate around Z (anteroposterior) axis | Shift + right drag (move horizontally) | Shift + Command + drag (move horizontally) |
| translate along X axis (holizontally) translate along Y axis (vertically) |
right drag | Command + drag |
| translate along Z axis (zoom in/out) | Shift + left drag | Shift + drag |
Operations such as changing display models and colors and saving the image, can be executed with a menu. The location of the menu is at the top of the application.
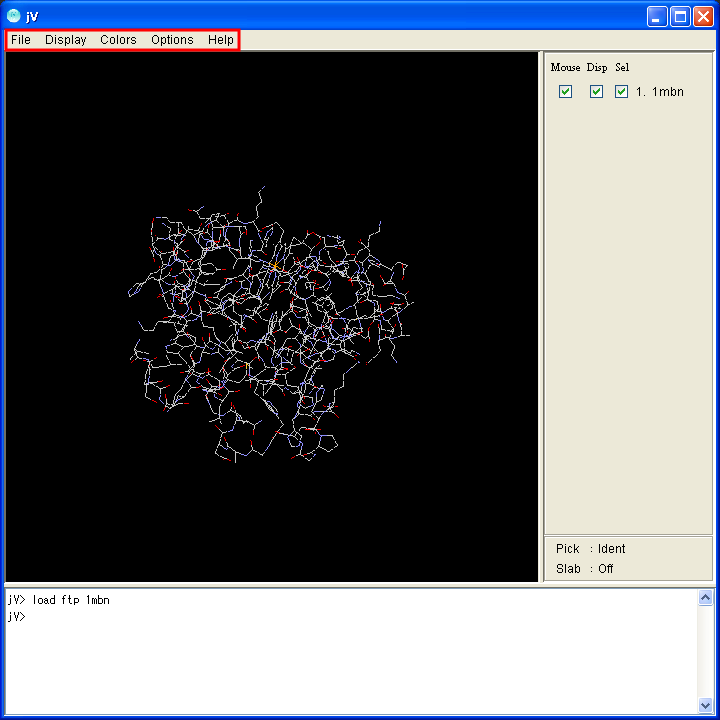 |
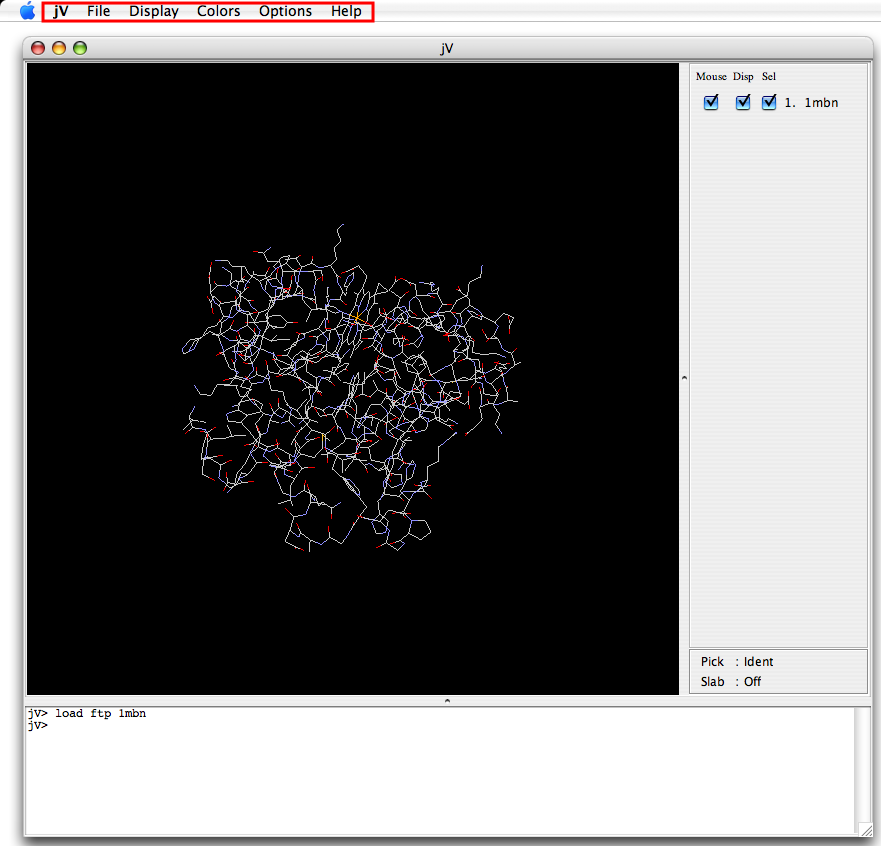 |
| Application window for Windows | Application window for Mac |
Basic menu operations are following (see also jV wiki for details):
| Operations | Menu location |
|---|---|
| Save a static image | [File]-[Save]-[PNG/JPEG] |
| Change the expression format | [File]-[Display]-[Wireframe/Backbone/Sticks/Spacefill/Ball&Stick/Ribbons/Cartoon] |
| Change the color | [File]-[Colors]-[Monochrome/CPK/Shapely/Group/Chain/Temperature/Structure/Charge/Amino] |
To execute more complicated operation such as diplaying some piece of the molecule or changing some part of molecule in another style, use command line operation.
See also the following links for details:
To run the jV application, it is necessary to install Java Runtime Environment (JRE) in advance. Please access the Java website to download and install it.
We finished providing the web start version. You can download the binary version (executable JAR file) and source code from the "jV Download" button below.
The latest version is 4.5.10.
In case you are to applet settings, you may refer the jV wiki.
When you hit upon some troubles in installing or viewing molecules, refer jV wiki - Trouble Shooting (in English) or jV Usage manual - Trouble Shooting (in Japanese).
The jV program was developed by Kengo Kinoshita (Graduate School of Information Sciences, Tohoku University) and Haruki Nakamura (Institute for Protein Research (IPR), Osaka University) as one of activity of Protein Data Bank Japan (PDBj), with the support from JST-NBDC and Institute for Protein Research, Osaka University.
