5FKW
 
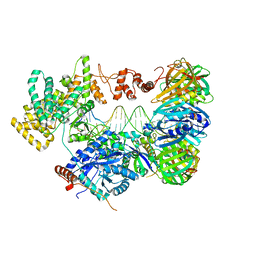 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5UOT
 
 | |
5FN2
 
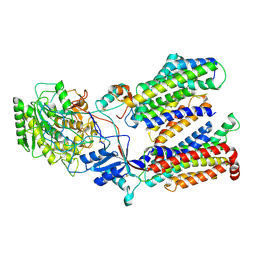 | | Cryo-EM structure of gamma secretase in complex with a drug DAPT | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
5FKU
 
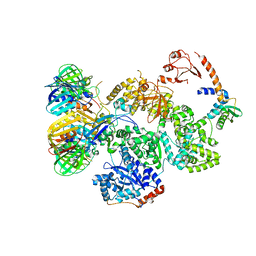 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FN3
 
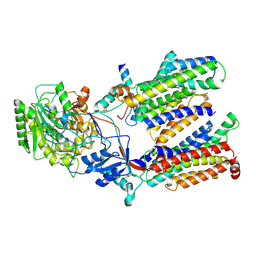 | | Cryo-EM structure of gamma secretase in class 1 of the apo- state ensemble | | Descriptor: | Gamma-secretase subunit APH-1A, Gamma-secretase subunit PEN-2, Nicastrin, ... | | Authors: | Bai, X.C, Rajendra, E, Yang, G.H, Shi, Y.G, Scheres, S.H.W. | | Deposit date: | 2015-11-10 | | Release date: | 2015-12-16 | | Last modified: | 2019-09-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Elife, 4, 2015
|
|
6ZHY
 
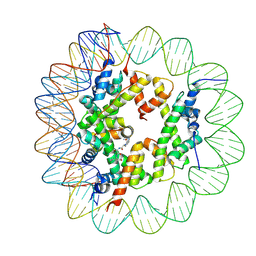 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
6ZHX
 
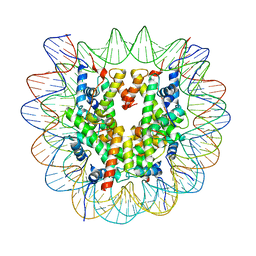 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: nucleosome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (145-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Croll, T.I, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
3J7P
 
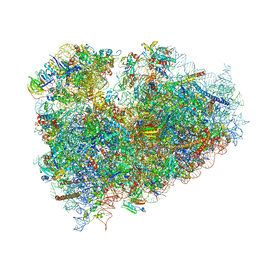 | | Structure of the 80S mammalian ribosome bound to eEF2 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J79
 
 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, large subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
3J9K
 
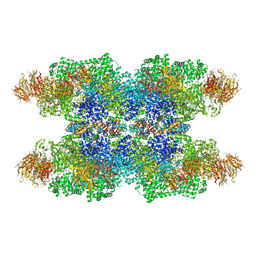 | | Structure of Dark apoptosome in complex with Dronc CARD domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apaf-1 related killer DARK, Caspase Nc | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
3J7R
 
 | | Structure of the translating mammalian ribosome-Sec61 complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J7Y
 
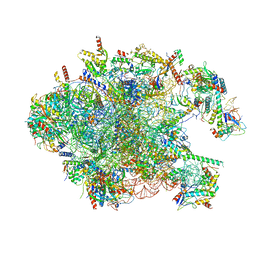 | | Structure of the large ribosomal subunit from human mitochondria | | Descriptor: | 16S rRNA, ADENOSINE MONOPHOSPHATE, CRIF1, ... | | Authors: | Brown, A, Amunts, A, Bai, X.C, Sugimoto, Y, Edwards, P.C, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the large ribosomal subunit from human mitochondria.
Science, 346, 2014
|
|
3J9L
 
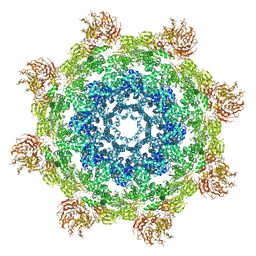 | | Structure of Dark apoptosome from Drosophila melanogaster | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
3J6B
 
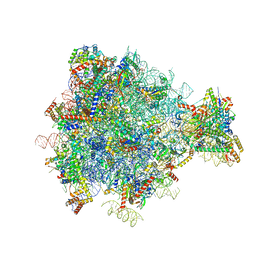 | | Structure of the yeast mitochondrial large ribosomal subunit | | Descriptor: | 21S ribosomal RNA, 54S ribosomal protein IMG1, mitochondrial, ... | | Authors: | Amunts, A, Brown, A, Bai, X.C, Llacer, J.L, Hussain, T, Emsley, P, Long, F, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the yeast mitochondrial large ribosomal subunit.
Science, 343, 2014
|
|
3J7O
 
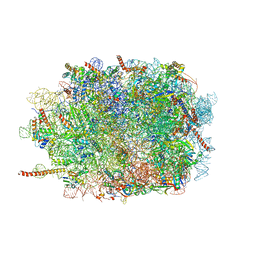 | | Structure of the mammalian 60S ribosomal subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J8H
 
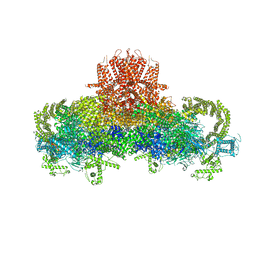 | | Structure of the rabbit ryanodine receptor RyR1 in complex with FKBP12 at 3.8 Angstrom resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Yan, Z, Bai, X, Yan, C, Wu, J, Scheres, S.H.W, Shi, Y, Yan, N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution.
Nature, 517, 2015
|
|
3J7A
 
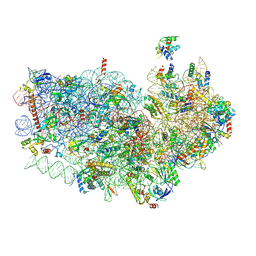 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, small subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
3J7Q
 
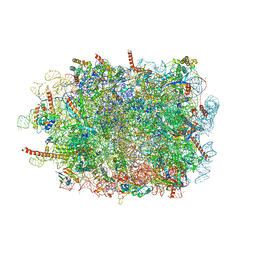 | | Structure of the idle mammalian ribosome-Sec61 complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
5WVE
 
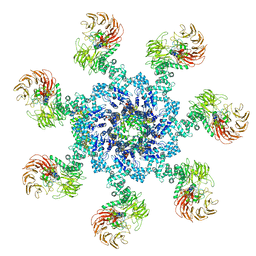 | | Apaf-1-Caspase-9 holoenzyme | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Caspase, ... | | Authors: | Li, Y, Zhou, M, Hu, Q, Shi, Y. | | Deposit date: | 2016-12-24 | | Release date: | 2017-02-08 | | Last modified: | 2017-03-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6VI3
 
 | | Straight Filament from Alzheimer's Disease Human Brain Tissue | | Descriptor: | GLYCINE, Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains
Cell, 180, 2020
|
|
7ARQ
 
 | | Cryo EM of 3D DNA origami 16 helix bundle | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Feigl, E, Kube, M, Kohler, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVX
 
 | | Cryo-EM structure of yeast CENP-OPQU+ bound to the CENP-A N-terminus | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CTF19, Inner kinetochore subunit MCM21, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
