7NNH
 
 | |
6QVD
 
 | |
6QV6
 
 | |
6QVB
 
 | |
6QVU
 
 | |
6QVC
 
 | |
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
3FD5
 
 | |
7MSV
 
 | |
4UMV
 
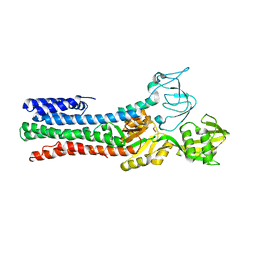 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN THE E2P STATE | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
4UMW
 
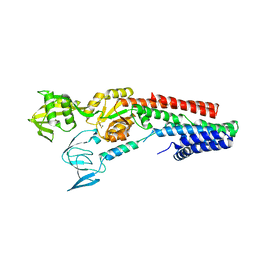 | | CRYSTAL STRUCTURE OF A ZINC-TRANSPORTING PIB-TYPE ATPASE IN E2.PI STATE | | Descriptor: | MAGNESIUM ION, TETRAFLUOROALUMINATE ION, ZINC-TRANSPORTING ATPASE | | Authors: | Wang, K.T, Sitsel, O, Meloni, G, Autzen, H.E, Andersson, M, Klymchuk, T, Nielsen, A.M, Rees, D.C, Nissen, P, Gourdon, P. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Structure and Mechanism of Zn(2+)-Transporting P-Type Atpases.
Nature, 514, 2014
|
|
3FD6
 
 | |
6U39
 
 | |
6DAF
 
 | | 2.4 Angstrom crystal structure of the F141L Ca/CaM:CaV1.2 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DAE
 
 | |
6DAD
 
 | | 1.65 Angstrom crystal structure of the N97I Ca/CaM:CaV1.2 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DAH
 
 | | 2.5 Angstrom crystal structure of the N97S CaM mutant | | Descriptor: | CALCIUM ION, Calmodulin-1 | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3N3Y
 
 | | Crystal structure of Thymidylate Synthase X (ThyX) from Helicobacter pylori with FAD and dUMP at 2.31A resolution | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase thyX | | Authors: | Wang, K, Wang, Q, Chen, J, Chen, L, Jiang, H, Shen, X. | | Deposit date: | 2010-05-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Structure, Enzymatic Characterization and Inhibitor Discovery of Thymidylate Synthase X (ThyX) from Helicobacter pylori Strain SS1
To be Published
|
|
8FAE
 
 | | Asymmetric structure of cleaved HIV-1 AE2 envelope glycoprotein trimer in styrene-maleic acid lipid nanoparticles (AE2.1) | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, K, Zhang, S, Sodroski, J, Mao, Y. | | Deposit date: | 2022-11-26 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles.
Commun Biol, 6, 2023
|
|
8FAD
 
 | | Asymmetric structure of cleaved HIV-1 AD8 envelope glycoprotein trimer in styrene-maleic acid lipid nanoparticles | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, K, Zhang, S, Sodroski, J, Mao, Y. | | Deposit date: | 2022-11-26 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles.
Commun Biol, 6, 2023
|
|
6V0L
 
 | | PDGFR-b Promoter Forms a G-Vacancy Quadruplex that Can be Complemented by dGMP: Molecular Structure and Recognition of Guanine Derivatives and Metabolites | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*(3D1)P*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Wang, K.B, Dickerhoff, J, Wu, G, Yang, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | SOLUTION NMR | | Cite: | PDGFR-beta Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs.
J.Am.Chem.Soc., 142, 2020
|
|
6U3D
 
 | |
7B52
 
 | | VAR2CSA full ectodomain | | Descriptor: | Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Gourdon, P.E, Dagil, R, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
6U3A
 
 | |
7B54
 
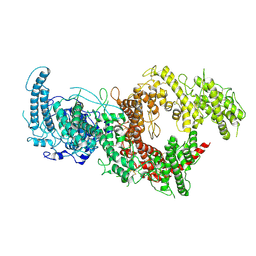 | | VAR2CSA full ectodomain in present of plCS, DBL1-DBL4 | | Descriptor: | VAR2CSA in presence of plCS, DBl1-DBL4,Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Dagil, R, Gourdon, P.E, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-06-02 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
