1ASH
 
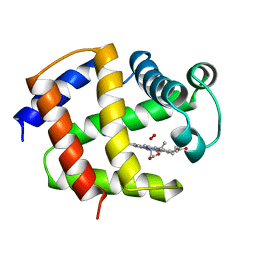 | | THE STRUCTURE OF ASCARIS HEMOGLOBIN DOMAIN I AT 2.2 ANGSTROMS RESOLUTION: MOLECULAR FEATURES OF OXYGEN AVIDITY | | Descriptor: | HEMOGLOBIN (OXY), OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, J, Mathews, F.S, Kloek, A.P, Goldberg, D.E. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of Ascaris hemoglobin domain I at 2.2 A resolution: molecular features of oxygen avidity.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
8W7F
 
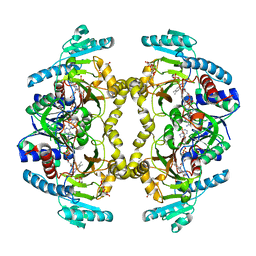 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase bound with FAD and a sulfate ion | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W78
 
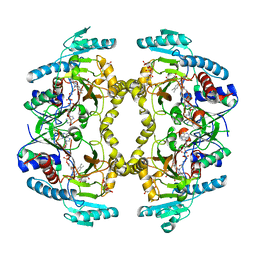 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase in complex with FAD and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DODECYL-BETA-D-MALTOSIDE, FI05204p, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W75
 
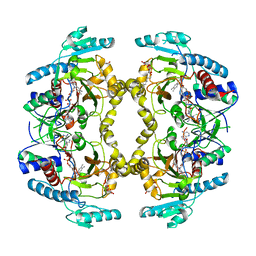 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
1FPR
 
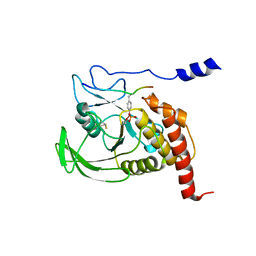 | | CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN THE CATALYTIC DOMAIN OF SHP-1 AND AN IN VITRO PEPTIDE SUBSTRATE PY469 DERIVED FROM SHPS-1. | | Descriptor: | PEPTIDE PY469, PROTEIN-TYROSINE PHOSPHATASE 1C | | Authors: | Yang, J, Cheng, Z, Niu, Z, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2000-08-31 | | Release date: | 2001-03-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 275, 2000
|
|
7N1R
 
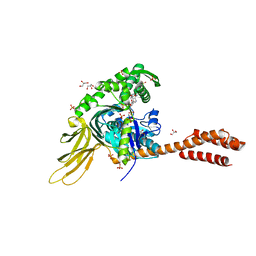 | | A novel and unique ATP hydrolysis to AMP by a human Hsp70 BiP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, Endoplasmic reticulum chaperone BiP, ... | | Authors: | Yang, J, Musayev, F, Liu, Q. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel and unique ATP hydrolysis to AMP by a human Hsp70 Binding immunoglobin protein (BiP).
Protein Sci., 31, 2022
|
|
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | Authors: | Yang, J, Lin, S, Lu, G.W. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
1PX8
 
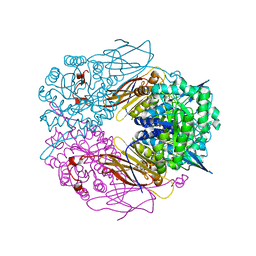 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
8XBD
 
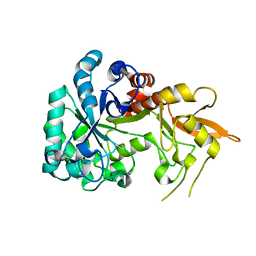 | |
8XHO
 
 | |
6VMG
 
 | | Chloroplast ATP synthase (O3, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.46 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
5IFG
 
 | | Crystal structure of HigA-HigB complex from E. Coli | | Descriptor: | Antitoxin HigA, mRNA interferase HigB | | Authors: | Yang, J.S, Zhou, K, Gao, z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural insight into the E. coli HigBA complex
Biochem. Biophys. Res. Commun., 478, 2016
|
|
2WMN
 
 | | Structure of the complex between DOCK9 and Cdc42-GDP. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2WM9
 
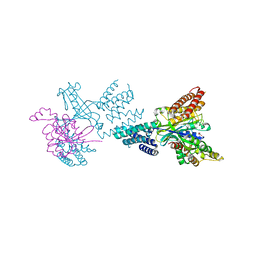 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GLYCEROL | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-22 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2WMO
 
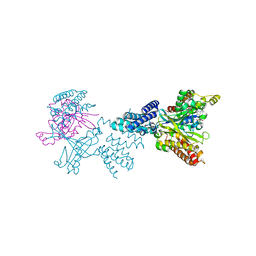 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2ASI
 
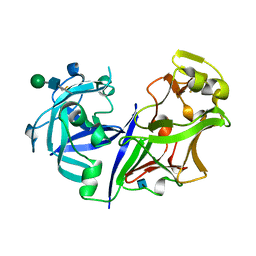 | | ASPARTIC PROTEINASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARTIC PROTEINASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Jia, Z, Vandonselaar, M, Kepliakov, P.S.A, Quail, J.W. | | Deposit date: | 1995-12-09 | | Release date: | 1996-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the aspartic proteinase from Rhizomucor miehei at 2.15 A resolution.
J.Mol.Biol., 268, 1997
|
|
8H5T
 
 | |
6VMB
 
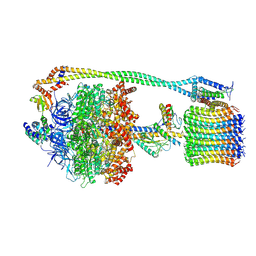 | | Chloroplast ATP synthase (C1, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (5.23 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VOG
 
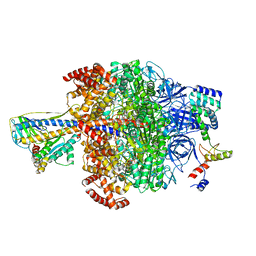 | | Chloroplast ATP synthase (O2, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VOJ
 
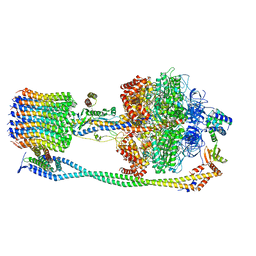 | | Chloroplast ATP synthase (R3, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VM1
 
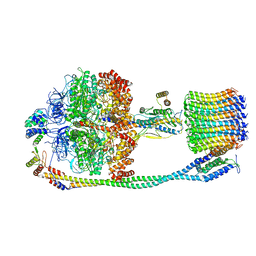 | | Chloroplast ATP synthase (C3, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VOM
 
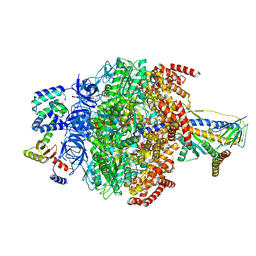 | | Chloroplast ATP synthase (R2, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VMD
 
 | | Chloroplast ATP synthase (C1, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VOO
 
 | | Chloroplast ATP synthase (R1, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VOK
 
 | | Chloroplast ATP synthase (R3, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
