6RNT
 
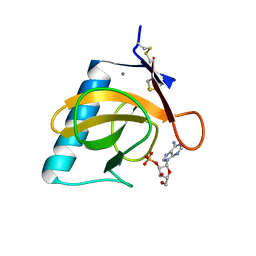 | | CRYSTAL STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH ADENOSINE 2'-MONOPHOSPHATE AT 1.8-ANGSTROMS RESOLUTION | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Ding, J, Koellner, G, Grunert, H.-P, Saenger, W. | | Deposit date: | 1991-08-20 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of ribonuclease T1 complexed with adenosine 2'-monophosphate at 1.8-A resolution.
J.Biol.Chem., 266, 1991
|
|
8YFJ
 
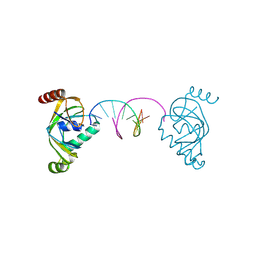 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-ATTGGATCCAAT-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*AP*TP*TP*GP*GP*AP*TP*CP*CP*AP*AP*T)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YEY
 
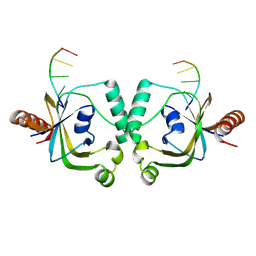 | | TRIP4 ASCH domain in complex with ssDNA-1 | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*TP*TP*TP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YEW
 
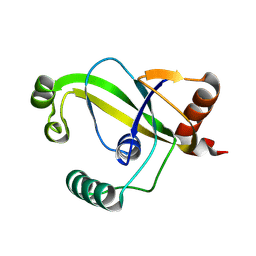 | | TRIP4 ASCH domain in unliganded form | | Descriptor: | Activating signal cointegrator 1 | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YFI
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCA-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YXX
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCG-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*CP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*G)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-04-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YXW
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCC-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-04-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
5B5R
 
 | | Crystal structure of GSDMA3 | | Descriptor: | Gasdermin-A3 | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-15 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Pore-forming activity and structural autoinhibition of the gasdermin family.
Nature, 535, 2016
|
|
1HNI
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN A COMPLEX WITH THE NONNUCLEOSIDE INHIBITOR ALPHA-APA R 95845 AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Ding, J, Das, K, Arnold, E. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution.
Structure, 3, 1995
|
|
1AVP
 
 | |
4YKC
 
 | |
4YKD
 
 | |
4YL6
 
 | |
8RNT
 
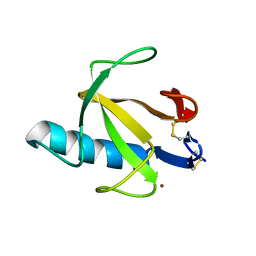 | | STRUCTURE OF RIBONUCLEASE T1 COMPLEXED WITH ZINC(II) AT 1.8 ANGSTROMS RESOLUTION: A ZN2+.6H2O.CARBOXYLATE CLATHRATE | | Descriptor: | RIBONUCLEASE T1, ZINC ION | | Authors: | Ding, J, Choe, H.-W, Granzin, J, Saenger, W. | | Deposit date: | 1991-09-23 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ribonuclease T1 complexed with zinc(II) at 1.8 A resolution: a Zn2+.6H2O.carboxylate clathrate.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1HPZ
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, POL POLYPROTEIN | | Authors: | Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
1HQE
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | POL POLYPROTEIN | | Authors: | Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 2000-12-15 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
8SA2
 
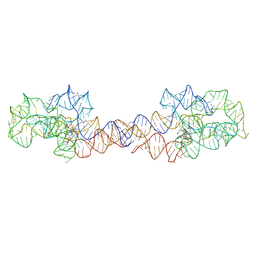 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA4
 
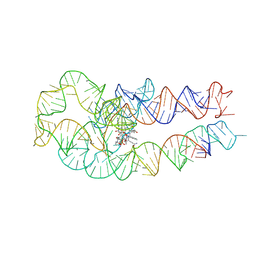 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA3
 
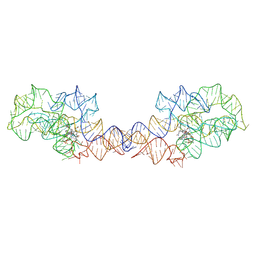 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
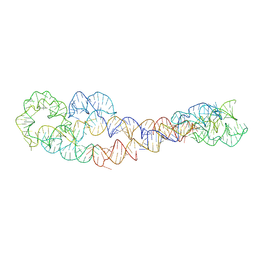 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
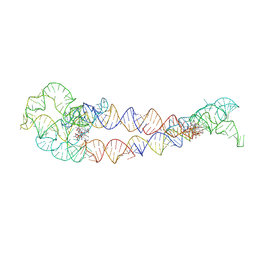 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
6KMV
 
 | |
6KN0
 
 | |
6KN1
 
 | | P20/P12 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-02 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KMZ
 
 | |
