8XR4
 
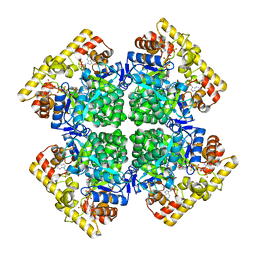 | | Crystal structure of AKRtyl-NADP(H) complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
8XR2
 
 | | Crystal structure of AKRtyl-apo1 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
8XR3
 
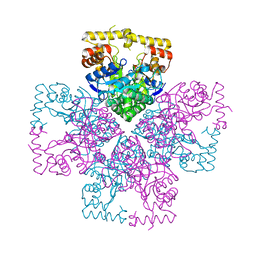 | | Crystal structure of AKRtyl-apo2 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D
Nat Commun, 15, 2024
|
|
5WVC
 
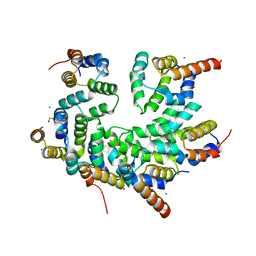 | | Structure of the CARD-CARD disk | | Descriptor: | Apoptotic protease-activating factor 1, Caspase, IODIDE ION | | Authors: | Lin, S.C, Lo, Y.C, Su, T.W. | | Deposit date: | 2016-12-24 | | Release date: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Structural Insights into DD-Fold Assembly and Caspase-9 Activation by the Apaf-1 Apoptosome.
Structure, 25, 2017
|
|
1PP0
 
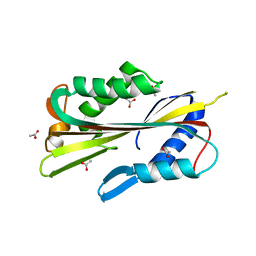 | | volvatoxin A2 in monoclinic crystal | | Descriptor: | ACETIC ACID, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2003-06-16 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
1PP6
 
 | |
8JBQ
 
 | |
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | Descriptor: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | Authors: | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
8JWM
 
 | | Crystal structure of AKRtyl-NADP-tylosin complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TYLOSIN | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWL
 
 | |
8JWO
 
 | |
8JWK
 
 | | The second purified state crystal structure of AKRtyl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
1VGF
 
 | | volvatoxin A2 (diamond crystal form) | | Descriptor: | ACETATE ION, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2004-04-24 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
1VCY
 
 | | VVA2 isoform | | Descriptor: | MALONATE ION, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2004-03-17 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
4A01
 
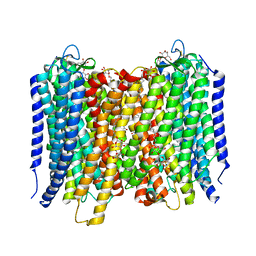 | | Crystal Structure of the H-Translocating Pyrophosphatase | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, IMIDODIPHOSPHORIC ACID, MAGNESIUM ION, ... | | Authors: | Lin, S.-M, Tsai, J.-Y, Hsiao, C.-D, Chiu, C.-L, Pan, R.-L, Sun, Y.-J. | | Deposit date: | 2011-09-07 | | Release date: | 2012-03-28 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of a Membrane Embedded H1-Translocating Pyrophosphatase
Nature, 484, 2012
|
|
7E9G
 
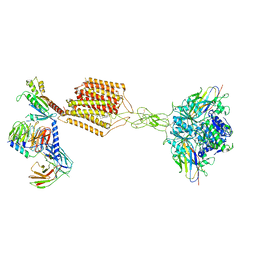 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7E9H
 
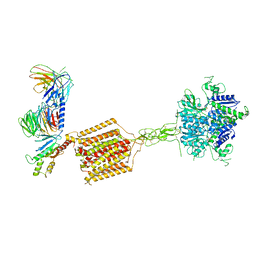 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
2POI
 
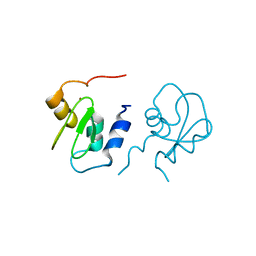 | | Crystal structure of XIAP BIR1 domain (I222 form) | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Lin, S. | | Deposit date: | 2007-04-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | XIAP Induces NF-kappaB Activation via the BIR1/TAB1 Interaction and BIR1 Dimerization.
Mol.Cell, 26, 2007
|
|
2QRA
 
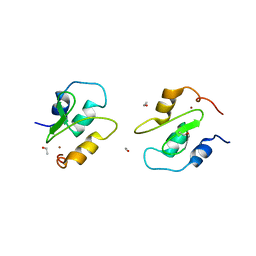 | | Crystal structure of XIAP BIR1 domain (P21 form) | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ETHANOL, ZINC ION | | Authors: | Lin, S.-C. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the BIR1 domain of XIAP in two crystal forms
J.Mol.Biol., 372, 2007
|
|
3MOP
 
 | | The ternary Death Domain complex of MyD88, IRAK4, and IRAK2 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, Interleukin-1 receptor-associated kinase-like 2, Myeloid differentiation primary response protein MyD88 | | Authors: | Lin, S.-C, Lo, Y.-C, Wu, H. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling.
Nature, 465, 2010
|
|
8JWN
 
 | | Crystal structure of AKRtyl-NADPH complex | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
1VF3
 
 | | cGSTA1-1 in complex with glutathione conjugate of CDNB | | Descriptor: | ACETIC ACID, GLUTATHIONE S-(2,4 DINITROBENZENE), Glutathione S-transferase 3 | | Authors: | Lin, S.C, Lo, Y.C, Tam, M.F, Liaw, Y.C. | | Deposit date: | 2004-04-07 | | Release date: | 2005-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of chicken glutathione S-transferase A1-1
To be Published
|
|
1VF2
 
 | |
