2QLK
 
 | | Adenovirus AD35 fibre head | | Descriptor: | Fiber, GLYCEROL | | Authors: | Liaw, Y.-C, Amiraslanov, I, Wang, H, Lieber, A. | | Deposit date: | 2007-07-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob
J.Virol., 81, 2007
|
|
4XWN
 
 | |
4XWL
 
 | | Catalytic domain of Clostridium Cellulovorans Exgs | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CALCIUM ION, ... | | Authors: | liaw, Y.-C. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structures of exoglucanase from Clostridium cellulovorans: cellotetraose binding and cleavage
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XWM
 
 | |
1ABR
 
 | | CRYSTAL STRUCTURE OF ABRIN-A | | Descriptor: | ABRIN-A, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Tahirov, T.H, Lu, T.-H, Liaw, Y.-C, Chu, S.-C, Lin, J.-Y. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of abrin-a at 2.14 A.
J.Mol.Biol., 250, 1995
|
|
1V2G
 
 | | The L109P mutant of E. coli Thioesterase I/Protease I/Lysophospholipase L1 (TAP) in complexed with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, IMIDAZOLE, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Lo, Y.-C, Lin, S.-C, Liaw, Y.-C. | | Deposit date: | 2003-10-15 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate specificities of Escherichia coli thioesterase I/protease I/lysophospholipase L1 are governed by its switch loop movement
Biochemistry, 44, 2005
|
|
1J00
 
 | |
1IVN
 
 | | E.coli Thioesterase I/Protease I/Lysophospholiase L1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioesterase I | | Authors: | Lo, Y.-C, Shaw, J.-F, Liaw, Y.-C. | | Deposit date: | 2002-03-27 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Escherichia coli Thioesterase I/Protease I/Lysophospholipase L1: Consensus Sequence Blocks Constitute the Catalytic Center of SGNH-hydrolases through a Conserved Hydrogen Bond Network
J.Mol.Biol., 330, 2003
|
|
1PP0
 
 | | volvatoxin A2 in monoclinic crystal | | Descriptor: | ACETIC ACID, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2003-06-16 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
1PP6
 
 | |
1U8U
 
 | | E. coli Thioesterase I/Protease I/Lysophospholiase L1 in complexed with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, GLYCEROL, OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Lo, Y.-C, Lin, S.-C, Liaw, Y.-C. | | Deposit date: | 2004-08-07 | | Release date: | 2005-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Substrate specificities of Escherichia coli thioesterase I/protease I/lysophospholipase L1 are governed by its switch loop movement
Biochemistry, 44, 2005
|
|
1VGF
 
 | | volvatoxin A2 (diamond crystal form) | | Descriptor: | ACETATE ION, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2004-04-24 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
1VCY
 
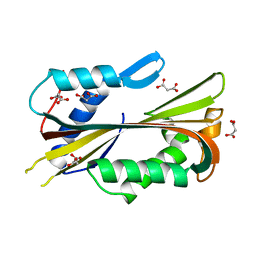 | | VVA2 isoform | | Descriptor: | MALONATE ION, volvatoxin A2 | | Authors: | Lin, S.-C, Lo, Y.-C, Lin, J.-Y, Liaw, Y.-C. | | Deposit date: | 2004-03-17 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures and electron micrographs of fungal volvatoxin A2
J.Mol.Biol., 343, 2004
|
|
1JRL
 
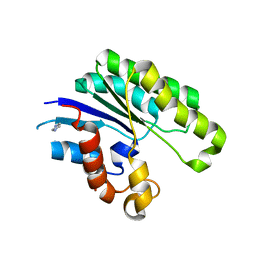 | | Crystal structure of E. coli Lysophospholiase L1/Acyl-CoA Thioesterase I/Protease I L109P mutant | | Descriptor: | Acyl-CoA Thioesterase I, IMIDAZOLE, SULFATE ION | | Authors: | Lo, Y.-C, Lin, S.-C, Shaw, J.-F, Liaw, Y.-C. | | Deposit date: | 2001-08-14 | | Release date: | 2003-07-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Escherichia coli Thioesterase I/Protease I/Lysophospholipase L1: Consensus Sequence Blocks Constitute the Catalytic Center of SGNH-hydrolases through a Conserved Hydrogen Bond Network
J.Mol.Biol., 330, 2003
|
|
1M07
 
 | | RESIDUES INVOLVED IN THE CATALYSIS AND BASE SPECIFICITY OF CYTOTOXIC RIBONUCLEASE FROM BULLFROG (RANA CATESBEIANA) | | Descriptor: | 5'-D(*AP*CP*GP*A)-3', Ribonuclease | | Authors: | Leu, Y.-J, Chern, S.-S, Wang, S.-C, Hsiao, Y.-Y, Amiraslanov, I, Liaw, Y.-C, Liao, Y.-D. | | Deposit date: | 2002-06-12 | | Release date: | 2003-01-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Residues involved in the catalysis, base specificity, and cytotoxicity of ribonuclease from Rana catesbeiana based upon mutagenesis and X-ray crystallography
J.Biol.Chem., 278, 2003
|
|
1D22
 
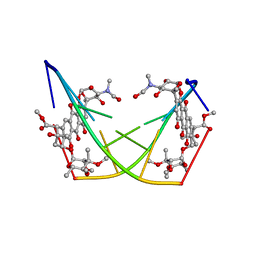 | | BINDING OF THE ANTITUMOR DRUG NOGALAMYCIN AND ITS DERIVATIVES TO DNA: STRUCTURAL COMPARISON | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*(AS)P*(5CM)P*G)-3'), U-58872, HYDROXY DERIVATIVE OF NOGALAMYCIN | | Authors: | Gao, Y.-G, Liaw, Y.-C, Robinson, H, Wang, A.H.-J. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of the antitumor drug nogalamycin and its derivatives to DNA: structural comparison.
Biochemistry, 29, 1990
|
|
1D21
 
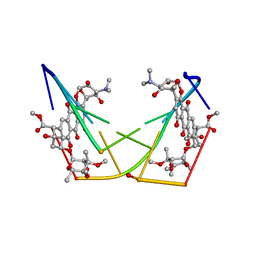 | | BINDING OF THE ANTITUMOR DRUG NOGALAMYCIN AND ITS DERIVATIVES TO DNA: STRUCTURAL COMPARISON | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*(AS)P*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Gao, Y.-G, Liaw, Y.-C, Robinson, H, Wang, A.H.-J. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of the antitumor drug nogalamycin and its derivatives to DNA: structural comparison.
Biochemistry, 29, 1990
|
|
1C8C
 
 | | CRYSTAL STRUCTURES OF THE CHROMOSOMAL PROTEINS SSO7D/SAC7D BOUND TO DNA CONTAINING T-G MISMATCHED BASE PAIRS | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', DNA-BINDING PROTEIN 7A | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Liaw, Y.-C, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 2000-05-04 | | Release date: | 2001-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs.
J.Mol.Biol., 303, 2000
|
|
1KM8
 
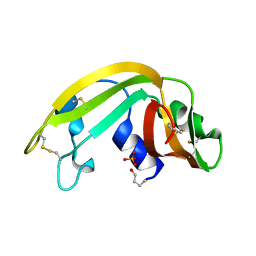 | | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog) | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE, OOCYTES | | Authors: | Chern, S.-S, Musayev, F.N, Amiraslanov, I.R, Liao, Y.-D, Liaw, Y.-C. | | Deposit date: | 2001-12-14 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog)
To be Published
|
|
1KM9
 
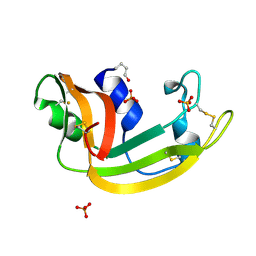 | | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog) | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE, OOCYTES | | Authors: | Chern, S.-S, Musayev, F.N, Amiraslanov, I.R, Liao, Y.-D, Liaw, Y.-C. | | Deposit date: | 2001-12-14 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Structure of a Cytotoxic Ribonuclease From the Oocyte of Rana Catesbeiana (Bullfrog)
To be Published
|
|
2D34
 
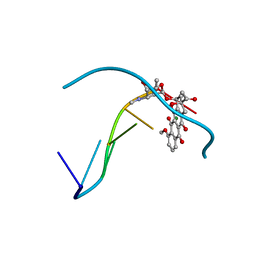 | | FORMALDEHYDE CROSS-LINKS DAUNORUBICIN AND DNA EFFICIENTLY: HPLC AND X-RAY DIFFRACTION STUDIES | | Descriptor: | 5'-D(*CP*GP*TP*(A35)P*CP*G)-3', DAUNOMYCIN, MAGNESIUM ION | | Authors: | Wang, A.H.-J, Gao, Y.-G, Liaw, Y.-C, Li, Y.-K. | | Deposit date: | 1991-05-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-ray diffraction studies.
Biochemistry, 30, 1991
|
|
1QPW
 
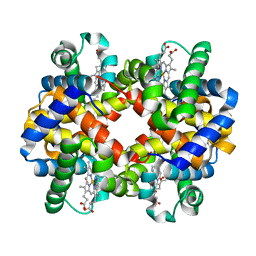 | | CRYSTAL STRUCTURE DETERMINATION OF PORCINE HEMOGLOBIN AT 1.8A RESOLUTION | | Descriptor: | OXYGEN MOLECULE, PORCINE HEMOGLOBIN (ALPHA SUBUNIT), PORCINE HEMOGLOBIN (BETA SUBUNIT), ... | | Authors: | Lu, T.-H, Panneerselvam, K, Liaw, Y.-C, Kan, P, Lee, C.-J. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of porcine haemoglobin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1D33
 
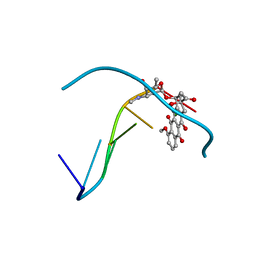 | | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-RAY diffraction studies | | Descriptor: | 5'-D(*CP*GP*CP*(G49)P*CP*G)-3', DAUNOMYCIN, MAGNESIUM ION | | Authors: | Wang, A.H.-J, Gao, Y.-G, Liaw, Y.-C, Li, Y.-K. | | Deposit date: | 1991-02-27 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-ray diffraction studies.
Biochemistry, 30, 1991
|
|
1D35
 
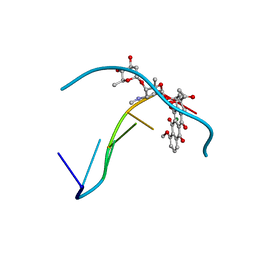 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(A40)P*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D36
 
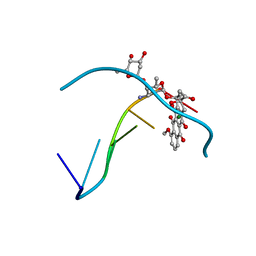 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
