1BMV
 
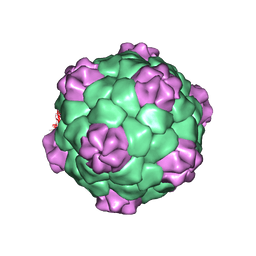 | | PROTEIN-RNA INTERACTIONS IN AN ICOSAHEDRAL VIRUS AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (ICOSAHEDRAL VIRUS - A DOMAIN), PROTEIN (ICOSAHEDRAL VIRUS - B AND C DOMAIN), RNA (5'-R(*GP*GP*UP*CP*AP*AP*AP*AP*UP*GP*C)-3') | | Authors: | Chen, Z, Stauffacher, C, Li, Y, Schmidt, T, Bomu, W, Kamer, G, Shanks, M, Lomonossoff, G, Johnson, J.E. | | Deposit date: | 1989-10-09 | | Release date: | 1989-10-09 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein-RNA interactions in an icosahedral virus at 3.0 A resolution.
Science, 245, 1989
|
|
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
1AHT
 
 | |
8SYM
 
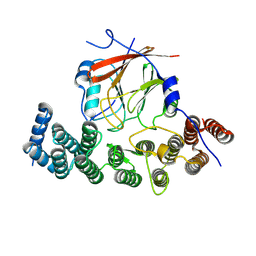 | | Human VPS29/VPS35L Complex (Locally refined map) | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 2023
|
|
8SYN
 
 | | Human VPS35L/VPS29/VPS26C Complex | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 2023
|
|
8SYO
 
 | | Human Retriever VPS35L/VPS29/VPS26C Complex (Composite Map) | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 2023
|
|
8TO0
 
 | | 48-nm repeating structure of doublets from mouse sperm flagella | | Descriptor: | Cilia- and flagella- associated protein 210, Cilia- and flagella-associated protein 107, Cilia- and flagella-associated protein 141, ... | | Authors: | Chen, Z, Shiozak, M, Hass, K.M, Skinner, W, Zhao, S, Guo, C, Polacco, B.J, Yu, Z, Krogan, N.J, Kaake, R.M, Vale, R.D, Agard, D.A. | | Deposit date: | 2023-08-02 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | De novo protein identification in mammalian sperm using in situ cryoelectron tomography and AlphaFold2 docking.
Cell, 186, 2023
|
|
5KU6
 
 | | Crystal structure for the complex of human carbonic anhydrase IV and methazolamide | | Descriptor: | ACETATE ION, Carbonic anhydrase 4, GLYCEROL, ... | | Authors: | Chen, Z, Waheed, A, Di Cera, E, Sly, W.S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intrinsic thermodynamics of high affinity inhibitor binding to recombinant human carbonic anhydrase IV.
Eur. Biophys. J., 47, 2018
|
|
4XOH
 
 | | Mechanistic insights into anchorage of the contractile ring from yeast to humans | | Descriptor: | Division mal foutue 1 protein | | Authors: | Chen, Z, Wu, J.-Q, Wang, J, Guan, R, Sun, L, Lee, I.-J, Liu, Y, Chen, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
7VZB
 
 | | Cryo-EM structure of C22:0-CoA bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] docosanethioate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7VWC
 
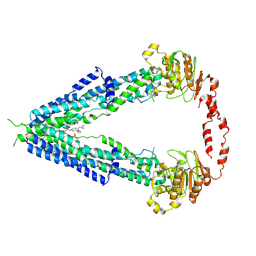 | | Cryo-EM structure of human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, [(2R)-3-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-oxidanyl-propyl] octadecanoate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-10 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7VX8
 
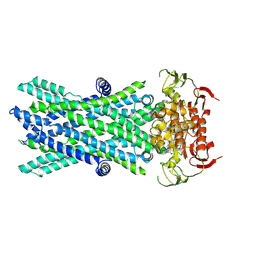 | | Cryo-EM structure of ATP-bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1 | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-12 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
5JXR
 
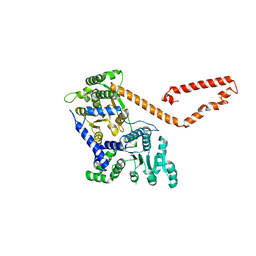 | | Crystal structure of MtISWI | | Descriptor: | CHLORIDE ION, Chromatin-remodeling complex ATPase-like protein | | Authors: | Chen, Z, Yan, L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structure and regulation of the chromatin remodeller ISWI
Nature, 540, 2016
|
|
1HSI
 
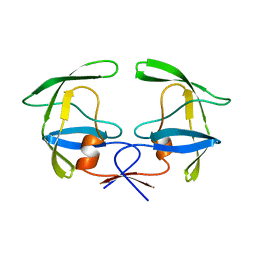 | |
1HSH
 
 | |
1HSG
 
 | |
5IXY
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 31: (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one | | Descriptor: | (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chen, Z, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
5NKP
 
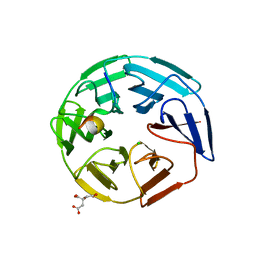 | | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK3 peptide | | Descriptor: | CHLORIDE ION, CITRIC ACID, Kelch-like protein 3, ... | | Authors: | Chen, Z, Sorrell, F.J, Pinkas, D.M, Williams, E, Mathea, S, Goubin, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Burgess-Brown, N, Bountra, C, Bullock, A. | | Deposit date: | 2017-03-31 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK3 peptide
To Be Published
|
|
6TTK
 
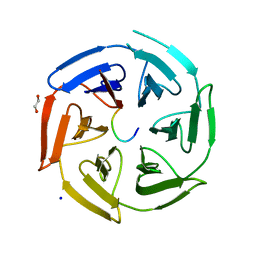 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
5J9U
 
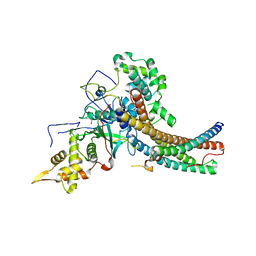 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9W
 
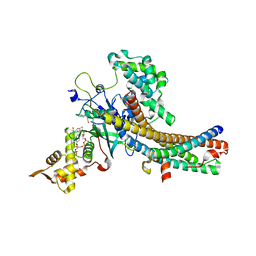 | | Crystal structure of the NuA4 core complex | | Descriptor: | ACETYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9T
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
5J9Q
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
1KV9
 
 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
1TOM
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, METHYL-PHE-PRO-AMINO-CYCLOHEXYLGLYCINE | | Authors: | Chen, Z. | | Deposit date: | 1996-12-06 | | Release date: | 1997-03-12 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, evaluation, and crystallographic analysis of L-371,912: A potent and selective active-site thrombin inhibitor.
Bioorg.Med.Chem.Lett., 7, 1997
|
|
