5X7D
 
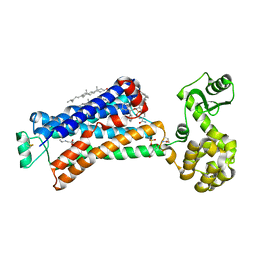 | | Structure of beta2 adrenoceptor bound to carazolol and an intracellular allosteric antagonist | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liu, X, Ahn, S, Kahsai, A.W, Meng, K.-C, Latorraca, N.R, Pani, B, Venkatakrishnan, A.J, Masoudi, A, Weis, W.I, Dror, R.O, Chen, X, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2017-02-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Mechanism of intracellular allosteric beta 2AR antagonist revealed by X-ray crystal structure.
Nature, 548, 2017
|
|
6W4S
 
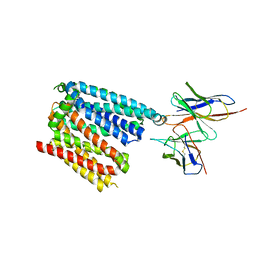 | | Structure of apo human ferroportin in lipid nanodisc | | Descriptor: | Fab45D8 Heavy Chain, Fab45D8 Light Chain, Solute carrier family 40 member 1 | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
5C1M
 
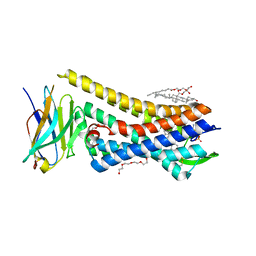 | | Crystal structure of active mu-opioid receptor bound to the agonist BU72 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3S,3aR,5aR,6R,11bR,11cS)-3a-methoxy-3,14-dimethyl-2-phenyl-2,3,3a,6,7,11c-hexahydro-1H-6,11b-(epiminoethano)-3,5a-methanonaphtho[2,1-g]indol-10-ol, CHOLESTEROL, ... | | Authors: | Huang, W.J, Manglik, A, Venkatakrishnan, A.J, Laeremans, T, Feinberg, E.N, Sanborn, A.L, Kato, H.E, Livingston, K.E, Thorsen, T.S, Kling, R, Granier, S, Gmeiner, P, Husbands, S.M, Traynor, J.R, Weis, W.I, Steyaert, J, Dror, R.O, Kobilka, B.K. | | Deposit date: | 2015-06-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insights into mu-opioid receptor activation.
Nature, 524, 2015
|
|
6WBV
 
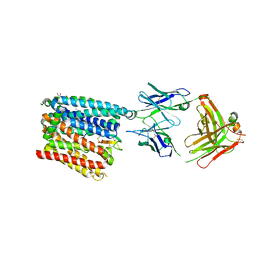 | | Structure of human ferroportin bound to hepcidin and cobalt in lipid nanodisc | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, COBALT (II) ION, Fab45D8 Heavy Chain, ... | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6W4V
 
 | | Structure of anti-ferroportin Fab45D8 | | Descriptor: | 1,2-ETHANEDIOL, Fab45D8 Heavy Chain, Fab45D8 Light Chain | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
4DAJ
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
8TA5
 
 | | Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA4
 
 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA2
 
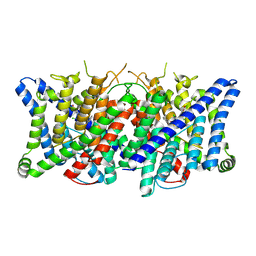 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42 | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA3
 
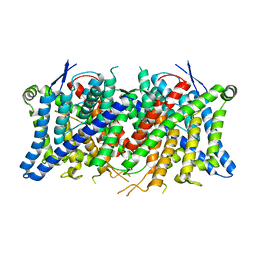 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA6
 
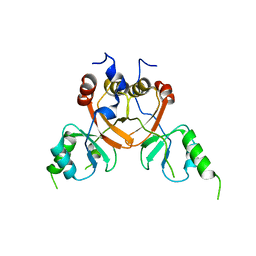 | | Cryo-EM structure of the human CLC-2 chloride channel C-terminal domain | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
5TVN
 
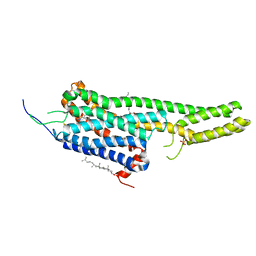 | | Crystal structure of the LSD-bound 5-HT2B receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, S, McCorvy, J.D, Betz, R.M, Venkatakrishnan, A.J, Levit, A, Lansu, K, Schools, Z.L, Che, T, Nichols, D.E, Shoichet, B.K, Dror, R.O, Roth, B.L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of an LSD-Bound Human Serotonin Receptor.
Cell, 168, 2017
|
|
5W0P
 
 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
5WIU
 
 | | Structure of the human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
5WIV
 
 | | Structure of the sodium-bound human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
7RMI
 
 | | SP6-11 biased agonist bound to active human neurokinin 1 receptor in complex with miniGs/q70 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
7RMG
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs/q70 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
7RMH
 
 | | Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Harris, J.A, Faust, B, Gondin, A.B, Daemgen, M.A, Suomivuori, C.M, Veldhuis, N.A, Cheng, Y, Dror, R.O, Thal, D, Manglik, A. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-03 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Selective G protein signaling driven by substance P-neurokinin receptor dynamics.
Nat.Chem.Biol., 18, 2022
|
|
6MXT
 
 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
6O3C
 
 | | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-4,7-difluoro-N-{[2-methoxy-5-(pyridin-4-yl)phenyl]methyl}-N-[trans-4-(methylamino)cyclohexyl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Deshpande, I.S, Liang, J, Hedeen, D, Roberts, K.J, Zhang, Y, Ha, B, Latorraca, N.R, Faust, B, Dror, R.O, Beachy, P.A, Myers, B.R, Manglik, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Smoothened stimulation by membrane sterols drives Hedgehog pathway activity.
Nature, 571, 2019
|
|
9BQJ
 
 | | RO76 bound muOR-Gi1-scFv16 complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Majumdar, S, Kobilka, B.K. | | Deposit date: | 2024-05-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling Modulation Mediated by Ligand Water Interactions with the Sodium Site at mu OR.
Acs Cent.Sci., 10, 2024
|
|
6PJX
 
 | | Crystal Structure of G Protein-Coupled Receptor Kinase 5 (GRK5) in Complex with Calmodulin (CaM) | | Descriptor: | CALCIUM ION, Calmodulin, G protein-coupled receptor kinase 5, ... | | Authors: | Bhardwaj, A, Komolov, K.E, Sulon, S, Benovic, J.L. | | Deposit date: | 2019-06-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a GRK5-Calmodulin Complex Reveals Molecular Mechanism of GRK Activation and Substrate Targeting.
Mol.Cell, 81, 2021
|
|
8SMV
 
 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Biorxiv, 2023
|
|
