3KHS
 
 | | Crystal structure of grouper iridovirus purine nucleoside phosphorylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Kang, Y.N, Zhang, Y, Allan, P.W, Parker, W.B, Ting, J.W, Chang, C.Y, Ealick, S.E. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of grouper iridovirus purine nucleoside phosphorylase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4YKQ
 
 | | Heat Shock Protein 90 Bound to CS301 | | Descriptor: | 1-(5-ethyl-2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS301
To Be Published
|
|
7KNX
 
 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNW
 
 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
4UIP
 
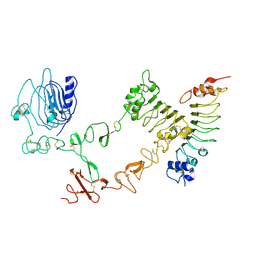 | | The complex structure of extracellular domain of EGFR with Repebody (rAC1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Kang, Y.J, Cha, Y.J, Cho, H.S, Lee, J.J, Kim, H.S. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Prenylation and Oxime Ligation for the Synthesis of Stable and Homogeneous Protein-Drug Conjugates for Targeted Therapy.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1WDS
 
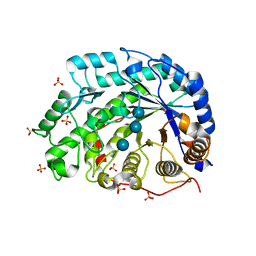 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDP
 
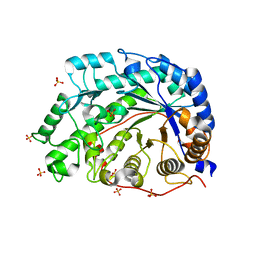 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDR
 
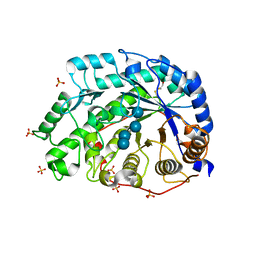 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDQ
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
3UAF
 
 | | Crystal Structure of a TTR-52 mutant of C. elegans | | Descriptor: | TTR-52 | | Authors: | Kang, Y.Y, Zhao, D.F, Liang, H.H, Liu, B, Liu, Q.W, Wang, X.C, Liu, Y.F. | | Deposit date: | 2011-10-21 | | Release date: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural study of TTR-52 reveals the mechanism by which a bridging molecule mediates apoptotic cell engulfment
Genes Dev., 26, 2012
|
|
8HJJ
 
 | | Anti-CRISPR protein AcrIC9 | | Descriptor: | Anti-CRISPR protein Type I-C9 | | Authors: | Kang, Y.J, Park, H.H. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of AcrIC9 revealing the putative inhibitory mechanism of AcrIC9 against the type IC CRISPR-Cas system.
Iucrj, 10, 2023
|
|
3QWY
 
 | | CED-2 | | Descriptor: | Cell death abnormality protein 2, GLYCEROL, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y, Sun, D, Hu, Y, Liu, Y.F. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
3SW7
 
 | |
3SW4
 
 | |
3QWX
 
 | | CED-2 1-174 | | Descriptor: | Cell death abnormality protein 2, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
3RN4
 
 | | Crystal structure of iron-substituted Sod2 from Saccharomyces cerevisiae | | Descriptor: | FE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Kang, Y, He, Y.-X, Cheng, W, Zhou, C.-Z, Li, W.-F. | | Deposit date: | 2011-04-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structures of native and Fe-substituted SOD2 from Saccharomyces cerevisiae
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4FW5
 
 | |
4FW7
 
 | |
1UKP
 
 | | Crystal structure of soybean beta-amylase mutant substituted at surface region | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Mikami, B, Utsumi, S. | | Deposit date: | 2003-08-31 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Change in the crystal packing of soybean beta-amylase mutants substituted at a few surface amino acid residues
Protein Eng., 16, 2003
|
|
1UKO
 
 | | Crystal structure of soybean beta-amylase mutant substituted at surface region | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Mikami, B, Utsumi, S. | | Deposit date: | 2003-08-30 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Change in the crystal packing of soybean beta-amylase mutants substituted at a few surface amino acid residues
Protein Eng., 16, 2003
|
|
4FW6
 
 | |
3JSK
 
 | | Thiazole synthase from Neurospora crassa | | Descriptor: | ADENOSINE DIPHOSPHATE 5-(BETA-ETHYL)-4-METHYL-THIAZOLE-2-CARBOXYLIC ACID, CyPBP37 protein, FE (II) ION | | Authors: | Kang, Y.N, Bale, S, Ealick, S.E. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of thiazole synthase Thi4 from Neurospora crassa
to be published
|
|
4EK5
 
 | |
4EK6
 
 | |
