6XKI
 
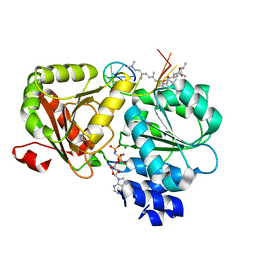 | | Crystal structure of eIF4A-I in complex with RNA bound to des-MePateA, a pateamine A analog | | Descriptor: | (3S,6Z,8E,11S,15R)-15-amino-3-[(1E,3E,5E)-7-(dimethylamino)-2,5-dimethylhepta-1,3,5-trien-1-yl]-9,11-dimethyl-4,12-dioxa-20-thia-21-azabicyclo[16.2.1]henicosa-1(21),6,8,18-tetraene-5,13-dione, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Liang, J, Naineni, S.K, Pelletier, J, Nagar, B. | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Functional mimicry revealed by the crystal structure of an eIF4A:RNA complex bound to the interfacial inhibitor, desmethyl pateamine A.
Cell Chem Biol, 28, 2021
|
|
1FPB
 
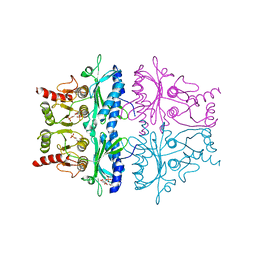 | | CRYSTAL STRUCTURE OF THE NEUTRAL FORM OF FRUCTOSE 1,6-BISPHOSPHATASE COMPLEXED WITH REGULATORY INHIBITOR FRUCTOSE 2,6-BISPHOSPHATE AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Liang, J.Y, Huang, S, Zhang, Y, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-02-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the neutral form of fructose 1,6-bisphosphatase complexed with regulatory inhibitor fructose 2,6-bisphosphate at 2.6-A resolution.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1CMI
 
 | | STRUCTURE OF THE HUMAN PIN/LC8 DIMER WITH A BOUND PEPTIDE | | Descriptor: | Dynein light chain 1, cytoplasmic, Nitric oxide synthase 1 | | Authors: | Liang, J, Guo, W, Jaffery, S, Snyder, S, Clardy, J. | | Deposit date: | 1999-05-06 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the PIN/LC8 dimer with a bound peptide.
Nat.Struct.Biol., 6, 1999
|
|
1NSG
 
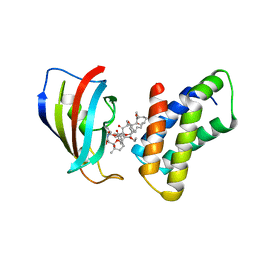 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FKBP-RAPAMYCIN ASSOCIATED PROTEIN (FRAP) | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1PHT
 
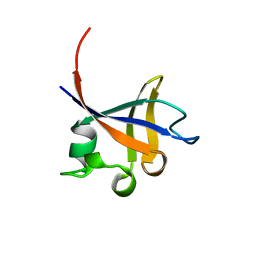 | | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN, RESIDUES 1-85 | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT | | Authors: | Liang, J, Chen, J.K, Schreiber, S.L, Clardy, J. | | Deposit date: | 1995-08-17 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of P13K SH3 domain at 20 angstroms resolution.
J.Mol.Biol., 257, 1996
|
|
3FAP
 
 | |
2FAP
 
 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-(C16)-ETHOXY RAPAMYCIN COMPLEX INTERACTING WITH HUMA | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FRAP | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1998-09-22 | | Release date: | 1999-05-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1PBK
 
 | | HOMOLOGOUS DOMAIN OF HUMAN FKBP25 | | Descriptor: | FKBP25, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Liang, J, Hung, D.T, Schreiber, S.L, Clardy, J. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the human 25 kDa FK506 binding protein complexed with rapamycin.
J.Am.Chem.Soc., 118, 1996
|
|
4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
8HPR
 
 | | LpqY-SugABC in state 4 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPS
 
 | | LpqY-SugABC in state 5 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
4FAP
 
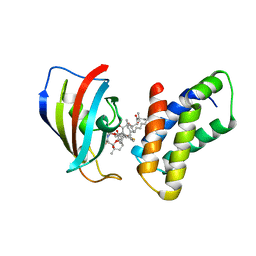 | |
8HPL
 
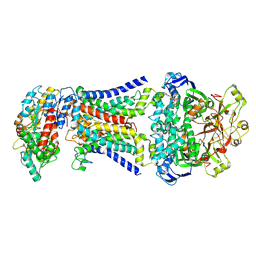 | | LpqY-SugABC in state 1 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPN
 
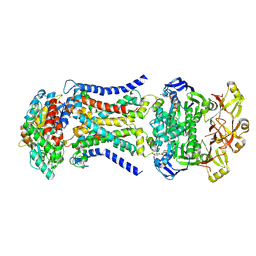 | | LpqY-SugABC in state 3 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPM
 
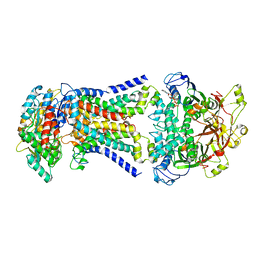 | | LpqY-SugABC in state 2 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
6PET
 
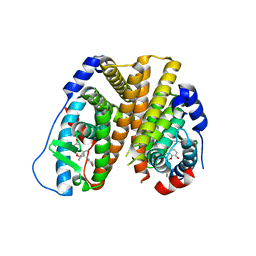 | | Crystal structure of 8-hydroxychromene compound 30 bound to estrogen receptor alpha | | Descriptor: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-8-ol, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, CHLORIDE ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Wang, X, Zbieg, J, Labadie, S.S, Zhang, B, Li, J, Liang, W. | | Deposit date: | 2019-06-20 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6PFM
 
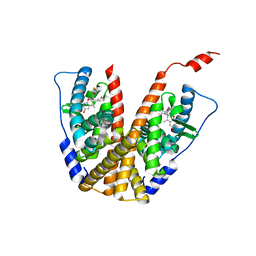 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | Descriptor: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7MSA
 
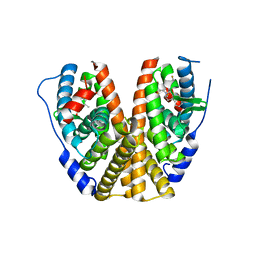 | | GDC-9545 in complex with estrogen receptor alpha | | Descriptor: | (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, 3-[(1R,3R)-1-(2,6-difluoro-4-{[1-(3-fluoropropyl)azetidin-3-yl]amino}phenyl)-3-methyl-1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl]-2,2-difluoropropan-1-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zbieg, J.R, Wang, X, Ortwine, D.F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-02 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | GDC-9545 (Giredestrant): A Potent and Orally Bioavailable Selective Estrogen Receptor Antagonist and Degrader with an Exceptional Preclinical Profile for ER+ Breast Cancer.
J.Med.Chem., 64, 2021
|
|
6DDT
 
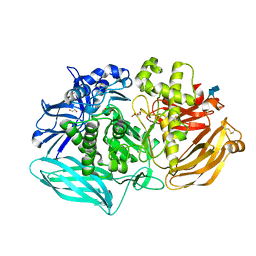 | | mouse beta-mannosidase (MANBA) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gytz, H, Liang, J, Liang, Y, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2018-05-10 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of mammalian beta-mannosidase provides insight into beta-mannosidosis and nystagmus.
FEBS J., 286, 2019
|
|
6DDU
 
 | | mouse beta-mannosidase bound to beta-D-mannose (MANBA) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gytz, H, Liang, J, Liang, Y, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2018-05-10 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.668 Å) | | Cite: | The structure of mammalian beta-mannosidase provides insight into beta-mannosidosis and nystagmus.
FEBS J., 286, 2019
|
|
1FRP
 
 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-2,6-BISPHOSPHATE, AMP AND ZN2+ AT 2.0 ANGSTROMS RESOLUTION. ASPECTS OF SYNERGISM BETWEEN INHIBITORS | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Xue, Y, Huang, S, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-08-26 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1FTA
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE, 1-PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH THE ALLOSTERIC INHIBITOR AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Lipscomb, W.N. | | Deposit date: | 1993-09-27 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric site of human liver fructose-1,6-bisphosphatase. Analysis of six AMP site mutants based on the crystal structure.
J.Biol.Chem., 269, 1994
|
|
6O3C
 
 | | Crystal structure of active Smoothened bound to SAG21k, cholesterol, and NbSmo8 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-4,7-difluoro-N-{[2-methoxy-5-(pyridin-4-yl)phenyl]methyl}-N-[trans-4-(methylamino)cyclohexyl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Deshpande, I.S, Liang, J, Hedeen, D, Roberts, K.J, Zhang, Y, Ha, B, Latorraca, N.R, Faust, B, Dror, R.O, Beachy, P.A, Myers, B.R, Manglik, A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Smoothened stimulation by membrane sterols drives Hedgehog pathway activity.
Nature, 571, 2019
|
|
6WOK
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 6 | | Descriptor: | (1R,3R)-1-(2,6-difluoro-4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Labadie, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Discovery of GNE-149 as a Full Antagonist and Efficient Degrader of Estrogen Receptor alpha for ER+ Breast Cancer.
Acs Med.Chem.Lett., 11, 2020
|
|
6UYA
 
 | | Crystal structure of Compound 19 bound to IRAK4 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{2-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-6-(morpholin-4-yl)-1-oxo-2,3-dihydro-1H-isoindol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Kiefer, J.R, Bryan, M.C, Lupardus, P.J, Zarrin, A.A, Rajapaksa, N.S, Gobbi, A, Drobnick, J, Kolesnikov, A, Liang, J, Do, S. | | Deposit date: | 2019-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Potent Benzolactam IRAK4 Inhibitors with Robust in Vivo Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
