4WIW
 
 | | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | Crystal structure of C-terminal domain of putative chitinase from Desulfitobacterium hafniense DCB-2
To Be Published
|
|
4WHI
 
 | | Crystal structure of C-terminal domain of penicillin binding protein Rv0907 | | Descriptor: | BROMIDE ION, Beta-lactamase, NICKEL (II) ION | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of C-terminal domain of penicillin binding protein Rv0907
To Be Published
|
|
1BC4
 
 | | THE SOLUTION STRUCTURE OF A CYTOTOXIC RIBONUCLEASE FROM THE OOCYTES OF RANA CATESBEIANA (BULLFROG), NMR, 15 STRUCTURES | | Descriptor: | RIBONUCLEASE | | Authors: | Chang, C.-F, Chen, C, Chen, Y.-C, Hom, K, Huang, R.-F, Huang, T. | | Deposit date: | 1998-05-05 | | Release date: | 1998-10-14 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a cytotoxic ribonuclease from the oocytes of Rana catesbeiana (bullfrog).
J.Mol.Biol., 283, 1998
|
|
4WER
 
 | | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Diacylglycerol kinase catalytic domain protein | | Authors: | Chang, C, Clancy, S, Hatzos-Skintges, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583
To Be Published
|
|
4W9R
 
 | | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271
To Be Published
|
|
4W66
 
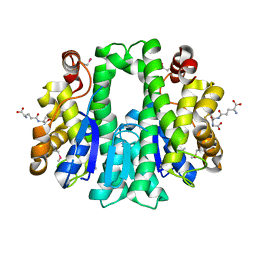 | |
8J9V
 
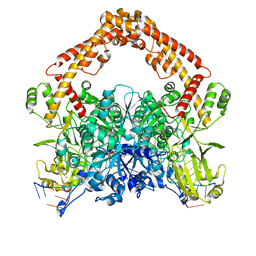 | | Cryo-EM structure of the African swine fever virus topoisomerase 2 complexed with Cut02aDNA and etoposide (EDI-1) | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*GP*AP*GP*GP*TP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*CP*GP*CP*CP*TP*AP*CP*AP*TP*AP*CP*CP*TP*C)-3'), ... | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
8TTP
 
 | | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-14 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of class C beta-lactamase from Escherichia coli in complex with avibactam
to be published
|
|
8J9W
 
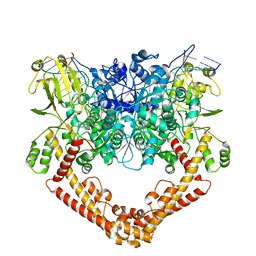 | | Cryo-EM structure of the African swine fever virus topoisomerase 2 complexed with Cut02bDNA and etoposide (EDI-2) | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*AP*AP*GP*AP*AP*CP*TP*CP*TP*GP*TP*AP*G)-3'), DNA (5'-D(*CP*AP*TP*GP*CP*TP*AP*CP*AP*GP*AP*GP*TP*TP*CP*TP*T)-3'), ... | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
8J9X
 
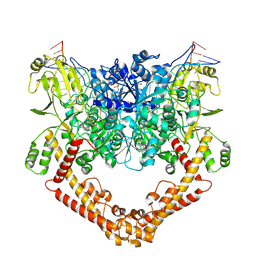 | |
8JA1
 
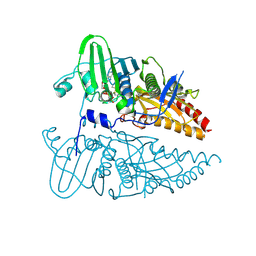 | |
8J8B
 
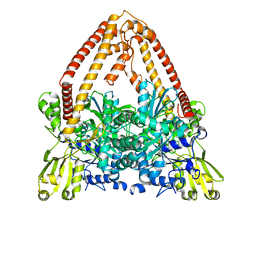 | |
8J8A
 
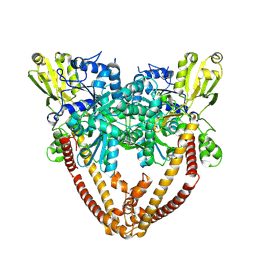 | |
8J89
 
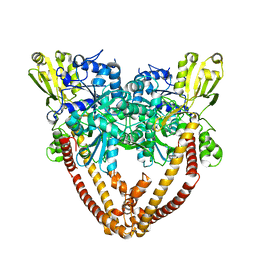 | |
8J8C
 
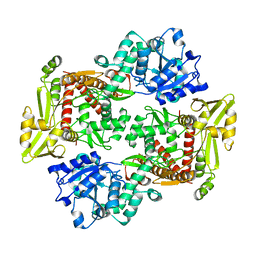 | |
8J88
 
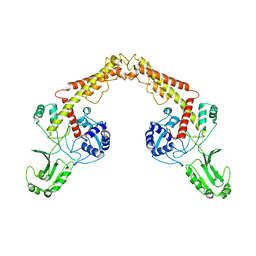 | | Asfv topoisomerase 2 - apo conformer Ib | | Descriptor: | DNA topoisomerase 2 | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
8J87
 
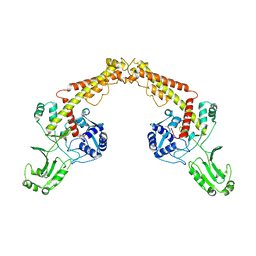 | | Asfv topoisomerase 2 - apo conformer Ia | | Descriptor: | DNA topoisomerase 2 | | Authors: | Chang, C.-W, Tsai, M.-D. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase.
Commun Chem, 7, 2024
|
|
8E86
 
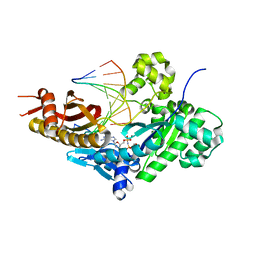 | | Human DNA polymerase eta-DNA-rC-ended primer-dGMPNPP ternary mismatch complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8K
 
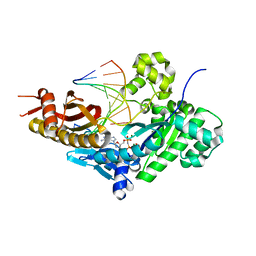 | | Human DNA polymerase eta-DNA-rC-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E87
 
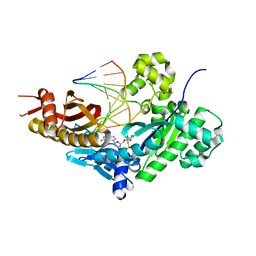 | | Human DNA polymerase eta-DNA-rA-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*TP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E89
 
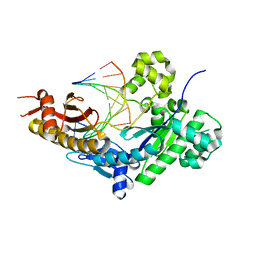 | | Human DNA polymerase eta-DNA-rU-ended primer-binary complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, DNA/RNA (5'-D(*AP*GP*CP*GP*TP*CP*A)-R(P*U)-3') | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8A
 
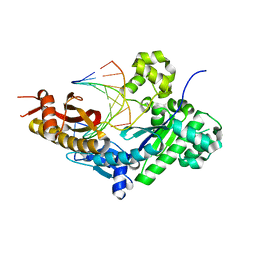 | | Human DNA polymerase eta-DNA-dT-ended primer binary complex | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E88
 
 | | Human DNA polymerase eta-DNA-rU-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8G
 
 | |
8E8E
 
 | |
