4E42
 
 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | CHLORIDE ION, NITRATE ION, SODIUM ION, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
3ULX
 
 | |
4PZG
 
 | | Crystal structure of human sorting nexin 10 (SNX10) | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, NITRATE ION, Sorting nexin-10 | | Authors: | Xu, T, Xu, J, Wang, Q, Liu, J. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human SNX10 reveals insights into its role in human autosomal recessive osteopetrosis.
Proteins, 82, 2014
|
|
4E41
 
 | | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Deng, L, Langley, R.J, Wang, Q, Topalian, S.L, Mariuzza, R.A. | | Deposit date: | 2012-03-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor G4
Proc.Natl.Acad.Sci.USA, 2012
|
|
4NRK
 
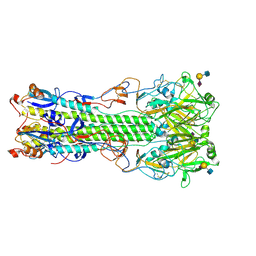 | | Structure of hemagglutinin with F95Y mutation of influenza virus B/Lee/40 complex with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Ni, F, Mbawuike, I.N, Kondrashkina, E, Wang, Q. | | Deposit date: | 2013-11-26 | | Release date: | 2014-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The roles of hemagglutinin Phe-95 in receptor binding and pathogenicity of influenza B virus.
Virology, 450-451, 2014
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
8X1W
 
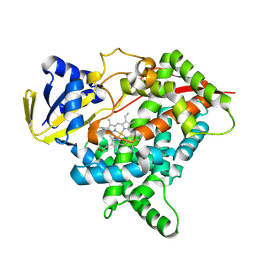 | | CYP725A4 apo structure | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Taxadiene 5-alpha hydroxylase | | Authors: | Chang, Z, Wang, Q. | | Deposit date: | 2023-11-09 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the Catalytic Mechanism of Taxadiene-5alpha-hydroxylase from Crystallography and Computational Analyses.
Acs Catalysis, 14, 2024
|
|
8X3E
 
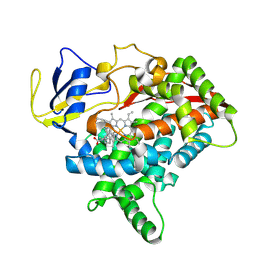 | | CYP725A4-Taxa-4,11-diene complex | | Descriptor: | (1~{R},3~{R},8~{R})-4,8,12,15,15-pentamethyltricyclo[9.3.1.0^{3,8}]pentadeca-4,11-diene, PROTOPORPHYRIN IX CONTAINING FE, Taxadiene 5-alpha hydroxylase | | Authors: | Chang, Z, Wang, Q. | | Deposit date: | 2023-11-13 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unraveling the Catalytic Mechanism of Taxadiene-5alpha-hydroxylase from Crystallography and Computational Analyses.
Acs Catalysis, 14, 2024
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
8HIT
 
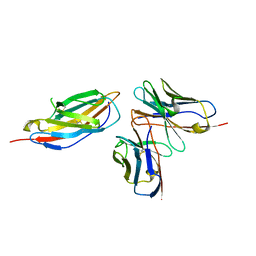 | | Crystal structure of anti-CTLA-4 humanized IgG1 MAb--JS007 in complex with human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, JS007-VH, JS007-VL | | Authors: | Tan, S, Shi, Y, Wang, Q, Gao, G.F, Guan, J, Chai, Y, Qi, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of the high-affinity anti-CTLA-4 monoclonal antibody JS007 for immune checkpoint therapy of cancer.
Mabs, 15, 2023
|
|
8H3D
 
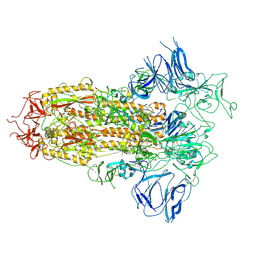 | | Structure of apo SARS-CoV-2 spike protein with one RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
8H3E
 
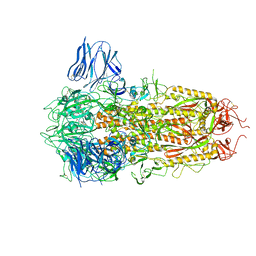 | | Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 spike protein, closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
7Y53
 
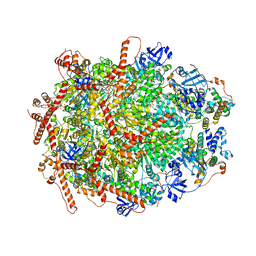 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
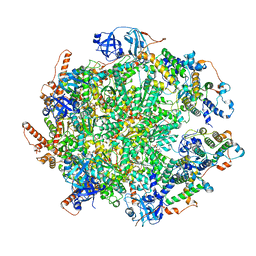 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
8XM1
 
 | | Phytase mutant APPAmut4 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phytase | | Authors: | Tu, T, Wang, Q. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mutant crystal structure of phytase APPAmut4 from Yersinia intermedia
To Be Published
|
|
2KDE
 
 | | NMR structure of major S5a (196-306):K48 linked diubiquitin species | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquitin | | Authors: | Zhang, N, Wang, Q, Ehlinger, A, Randles, L, Lary, J.W, Kang, Y, Haririnia, A, Cole, J.L, Fushman, D, Walters, K.J. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-01 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13.
Mol.Cell, 35, 2009
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7CXM
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
2PFD
 
 | | Anisotropically refined structure of FTCD | | Descriptor: | Formimidoyltransferase-cyclodeaminase | | Authors: | Poon, B.K, Chen, X, Lu, M, Quiocho, F.A, Wang, Q, Ma, J. | | Deposit date: | 2007-04-04 | | Release date: | 2007-04-24 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Anisotropically refined structure of FTCD
To be Published
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
