8WCT
 
 | |
1MHC
 
 | | MODEL OF MHC CLASS I H2-M3 WITH NONAPEPTIDE FROM RAT ND1 REFINED AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I ANTIGEN H2-M3, NONAPEPTIDE FROM RAT NADH DEHYDROGENASE | | Authors: | Wang, C.-R, Fischer Lindahl, K, Deisenhofer, J. | | Deposit date: | 1995-08-23 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3
Cell(Cambridge,Mass.), 82, 1995
|
|
4EVA
 
 | |
4QIN
 
 | | Structure of the human smoothened receptor in complex with SAG1.5 | | Descriptor: | 3-chloro-4,7-difluoro-N-[trans-4-(methylamino)cyclohexyl]-N-[3-(pyridin-4-yl)benzyl]-1-benzothiophene-2-carboxamide, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
4QIM
 
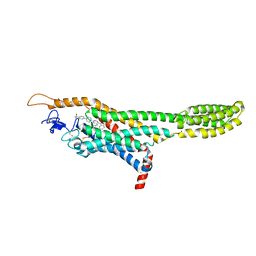 | | Structure of the human smoothened receptor in complex with ANTA XV | | Descriptor: | 2-{6-[4-(4-benzylphthalazin-1-yl)piperazin-1-yl]pyridin-3-yl}propan-2-ol, Smoothened homolog/Soluble cytochrome b562 chimeric protein, ZINC ION | | Authors: | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
8DSP
 
 | |
6UK1
 
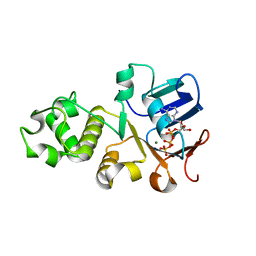 | | Crystal structure of nucleotide-binding domain 2 (NBD2) of the human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Vorobiev, S.M, Vernon, R.M, Khazanov, N, Senderowitz, H, Forman-Kay, J.D, Hunt, J.F. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | A thermodynamically stabilized form of the second nucleotide binding domain from human CFTR shows a catalytically inactive conformation
To Be Published
|
|
1P2X
 
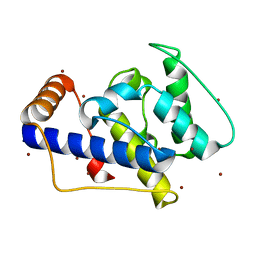 | | CRYSTAL STRUCTURE OF THE CALPONIN-HOMOLOGY DOMAIN OF RNG2 FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | BROMIDE ION, Ras GTPase-activating-like protein | | Authors: | Wang, C.-H, Balasubramanian, M.K, Dokland, T. | | Deposit date: | 2003-04-16 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure, crystal packing and molecular dynamics of the calponin-homology domain of Schizosaccharomyces pombe Rng2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8DMF
 
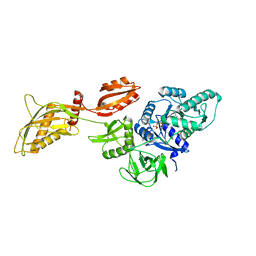 | | Cryo-EM structure of the ribosome-bound Bacteroides thetaiotaomicron EF-G2 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Tetracycline resistance protein TetQ | | Authors: | Wang, C, Han, W, Groisman, E.A, Liu, J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gut colonization by Bacteroides requires translation by an EF-G paralog lacking GTPase activity.
Embo J., 2022
|
|
2BHI
 
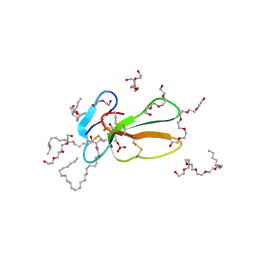 | | Crystal structure of Taiwan cobra cardiotoxin A3 complexed with sulfogalactoceramide | | Descriptor: | CYTOTOXIN 3, HEXAETHYLENE GLYCOL MONODECYL ETHER, SULFOGALACTOCERAMIDE | | Authors: | Wang, C.-H, Liu, J.-H, Wu, P.-L, Lee, S.-C, Hsiao, C.-D, Wu, W.-G. | | Deposit date: | 2005-01-12 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glycosphingolipid-Facilitated Membrane Insertion and Internalization of Cobra Cardiotoxin: The Sulfatide/Cardiotoxin Complex Structure in a Membrane-Like Environment Suggests a Lipid-Dependent Cell-Penetrating Mechanism for Membrane Binding Polypeptides.
J.Biol.Chem., 281, 2006
|
|
2V9C
 
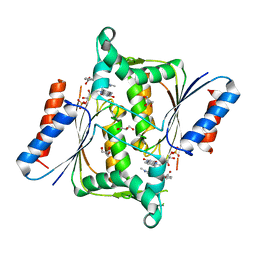 | | X-ray Crystallographic Structure of a Pseudomonas aeruginosa Azoreductase in Complex with Methyl Red. | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-DEPENDENT NADH-AZOREDUCTASE 1, ... | | Authors: | Wang, C.-J, Hagemeier, C, Rahman, N, Lowe, E.D, Noble, M.E.M, Coughtrie, M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-08-23 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular Cloning, Characterisation and Ligand- Bound Structure of an Azoreductase from Pseudomonas Aeruginosa
J.Mol.Biol., 373, 2007
|
|
4O9R
 
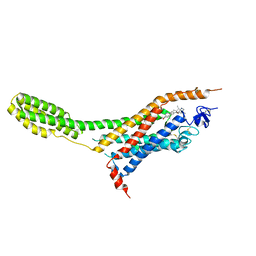 | | Human Smoothened Receptor structure in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Weierstall, U, James, D, White, T.A, Wang, D, Liu, W, Spence, J.C.H, Doak, R.B, Nelson, G, Fromme, P, Fromme, R, Grotjohann, I, Kupitz, C, Zatsepin, N.A, Liu, H, Basu, S, Wacker, D, Han, G.W, Katritch, V, Boutet, S, Messerschmidt, M, Willams, G.J, Koglin, J.E, Seibert, M.M, Klinker, M, Gati, C, Shoeman, R.L, Barty, A, Chapman, H.N, Kirian, R.A, Beyerlein, K.R, Stevens, R.C, Li, D, Shah, S.T.A, Howe, N, Caffrey, M, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography.
Nat Commun, 5, 2014
|
|
4JPZ
 
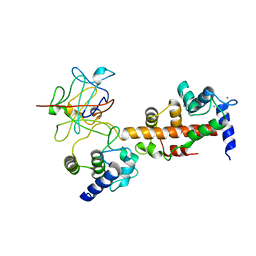 | | Voltage-gated sodium channel 1.2 C-terminal domain in complex with FGF13U and Ca2+/calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Fibroblast growth factor 13, ... | | Authors: | Wang, C, Chung, B.C, Yan, H, Wang, H.G, Lee, S.Y, Pitt, G.S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural analyses of Ca(2+)/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation.
Nat Commun, 5, 2014
|
|
4JQ0
 
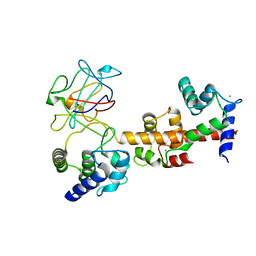 | | Voltage-gated sodium channel 1.5 C-terminal domain in complex with FGF12B and Ca2+/calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Fibroblast growth factor 12, ... | | Authors: | Wang, C, Chung, B.C, Yan, H, Wang, H.G, Lee, S.Y, Pitt, G.S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Structural analyses of Ca(2+)/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation.
Nat Commun, 5, 2014
|
|
4EL1
 
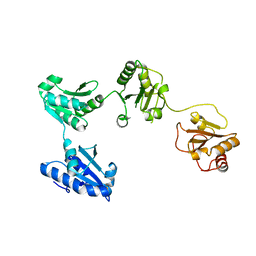 | | Crystal structure of oxidized hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4Y33
 
 | | Crystal of NO66 in complex with Ni(II)and N-oxalylglycine (NOG) | | Descriptor: | Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5INZ
 
 | | Racemic structure of baboon theta defensin-2 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Theta defensin-2, ... | | Authors: | Wang, C.K, King, G.J, Conibear, A.C, Ramos, M.C, Craik, D.J. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Mirror Images of Antimicrobial Peptides Provide Reflections on Their Functions and Amyloidogenic Properties.
J.Am.Chem.Soc., 138, 2016
|
|
1F0Z
 
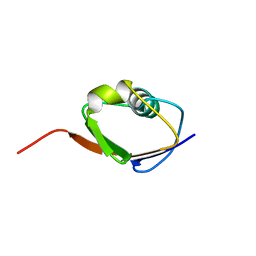 | | SOLUTION STRUCTURE OF THIS, THE SULFUR CARRIER PROTEIN IN E.COLI THIAMIN BIOSYNTHESIS | | Descriptor: | THIS PROTEIN | | Authors: | Wang, C, Xi, J, Begley, T.P, Nicholson, L.K. | | Deposit date: | 2000-05-17 | | Release date: | 2001-01-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ThiS and implications for the evolutionary roots of ubiquitin.
Nat.Struct.Biol., 8, 2001
|
|
4EKZ
 
 | | Crystal structure of reduced hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
6DL0
 
 | | Crystal structure of pohlianin C, an orbitide from Jatropha pohliana | | Descriptor: | pohlianin C | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
4Y3O
 
 | | Crystal structure of Ribosomal oxygenase NO66 in complex with substrate Rpl8 peptide and Ni(II) and cofactor N-oxalyglycine | | Descriptor: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, GLYCEROL, ... | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7UFJ
 
 | | Structure of human MR1-ethylvanillin in complex with human MAIT A-F7 TCR | | Descriptor: | 3-ethoxy-4-hydroxybenzaldehyde, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Wang, C.J, Rossjohn, J, Le Nours, J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative affinity measurement of small molecule ligand binding to Major Histocompatibility Complex class-I related protein 1 MR1.
J.Biol.Chem., 298, 2022
|
|
4H27
 
 | |
7D7N
 
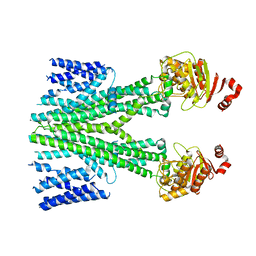 | | Cryo-EM structure of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
7D7R
 
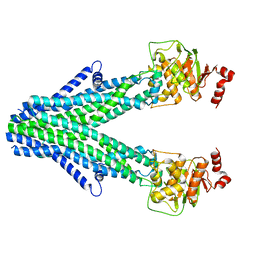 | | Cryo-EM structure of the core domain of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
