4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
2L3T
 
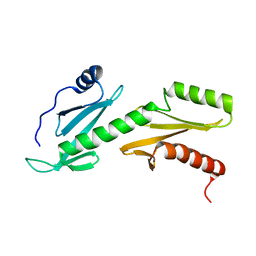 | | Solution structure of tandem SH2 domain from Spt6 | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Liu, J, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-09-22 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tandem SH2 domains from Spt6 and their binding to the phosphorylated RNA polymerase II C-terminal domain
To be Published
|
|
5WWD
 
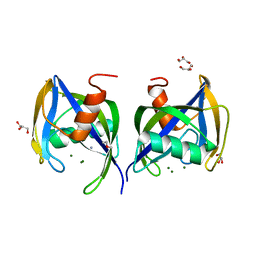 | | Crystal structure of AtNUDX1 | | Descriptor: | AMMONIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2016-12-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WY6
 
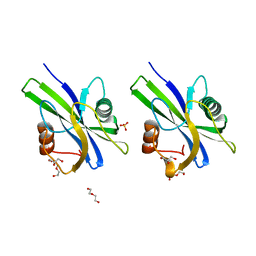 | | Crystal structure of AtNUDX1 (E56A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW7
 
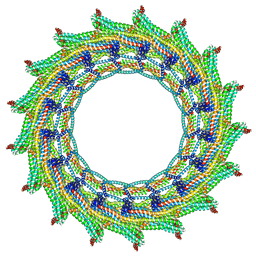 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
5X0W
 
 | | Molecular mechanism for the binding between Sharpin and HOIP | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Sharpin | | Authors: | Liu, J, Li, F, Cheng, X, Pan, L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into SHARPIN-Mediated Activation of HOIP for the Linear Ubiquitin Chain Assembly
Cell Rep, 21, 2017
|
|
6IR8
 
 | | Rice WRKY/DNA complex | | Descriptor: | DNA (5'-D(P*GP*AP*TP*AP*TP*TP*TP*GP*AP*CP*CP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*GP*TP*CP*AP*AP*AP*TP*AP*TP*C)-3'), OsWRKY45, ... | | Authors: | Liu, J, Cheng, X, Wang, D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain.
Nucleic Acids Res., 47, 2019
|
|
6V9C
 
 | | Crystal structure of FGFR4 kinase domain in complex with covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3R,4S)-4-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl]amino}oxolan-3-yl]prop-2-enamide, SULFATE ION | | Authors: | Liu, J, Liu, H. | | Deposit date: | 2019-12-13 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Selective, Covalent FGFR4 Inhibitors with Antitumor Activity in Models of Hepatocellular Carcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
6WXJ
 
 | | CSF1R signaling is a regulator of pathogenesis in progressive MS | | Descriptor: | 4-(3-{[(2S)-2-(6-methoxypyridin-3-yl)-2,3-dihydro-1,4-benzodioxin-6-yl]methyl}-3H-imidazo[4,5-b]pyridin-6-yl)-2-methylbut-3-yn-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Liu, J. | | Deposit date: | 2020-05-10 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | CSF1R signaling is a regulator of pathogenesis in progressive MS.
Cell Death Dis, 11, 2020
|
|
6PW1
 
 | | Cytochrome c Oxidase delta 16 | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, S. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural changes at the surface of cytochrome c oxidase alter the proton-pumping stoichiometry.
Biochim Biophys Acta Bioenerg, 1861, 2019
|
|
6PW0
 
 | | Cytochrome C oxidase delta 6 mutant | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, S. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural changes at the surface of cytochrome c oxidase alter the proton-pumping stoichiometry.
Biochim Biophys Acta Bioenerg, 1861, 2019
|
|
7RGW
 
 | |
3C2G
 
 | | Crystal complex of SYS-1/POP-1 at 2.5A resolution | | Descriptor: | Pop-1 8-residue peptide, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
3C2H
 
 | | Crystal Structure of SYS-1 at 2.6A resolution | | Descriptor: | CITRATE ANION, GLYCEROL, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
1AIL
 
 | | N-TERMINAL FRAGMENT OF NS1 PROTEIN FROM INFLUENZA A VIRUS | | Descriptor: | NONSTRUCTURAL PROTEIN NS1 | | Authors: | Liu, J, Lynch, P.A, Chien, C, Montelione, G.T, Krug, R.M, Berman, H.M. | | Deposit date: | 1997-04-21 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein.
Nat.Struct.Biol., 4, 1997
|
|
3ZP3
 
 | | INFLUENZA VIRUS (VN1194) H5 HA A138V mutant with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZP1
 
 | | INFLUENZA VIRUS (VN1194) H5 HA with LSTc | | Descriptor: | HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-2)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZP2
 
 | | INFLUENZA VIRUS (VN1194) H5 HA A138V mutant with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
8E7X
 
 | | RsTSPO A138F with one Heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7Z
 
 | | RsTSPO mutant -A138F | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
3ZP6
 
 | | INFLUENZA VIRUS (VN1194) H5 E190D mutant HA with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZP0
 
 | | INFLUENZA VIRUS (VN1194) H5 HA with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZPB
 
 | | INFLUENZA VIRUS (VN1194) H5 E190D mutant HA with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-27 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
