7EYD
 
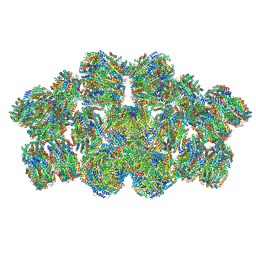 | | Cryo-EM structure of cyanobacterial phycobilisome from Anabaena sp. PCC 7120 | | Descriptor: | Allophycocyanin subunit alpha 1, Allophycocyanin subunit alpha-B, Allophycocyanin subunit beta, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-30 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
8HFQ
 
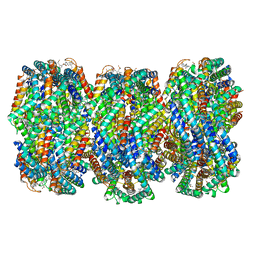 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | Authors: | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
7EXT
 
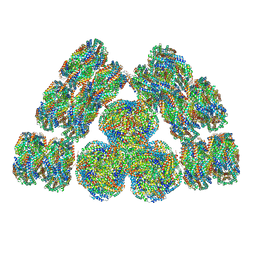 | | Cryo-EM structure of cyanobacterial phycobilisome from Synechococcus sp. PCC 7002 | | Descriptor: | Allophycocyanin alpha subunit, Allophycocyanin beta subunit, Allophycocyanin subunit alpha-B, ... | | Authors: | Zheng, L, Zheng, Z, Li, X, Wang, G, Zhang, K, Wei, P, Zhao, J, Gao, N. | | Deposit date: | 2021-05-28 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insight into the mechanism of energy transfer in cyanobacterial phycobilisomes.
Nat Commun, 12, 2021
|
|
1XQE
 
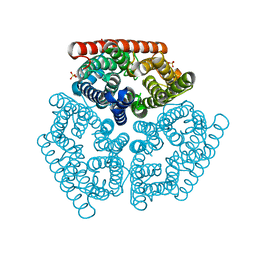 | | The mechanism of ammonia transport based on the crystal structure of AmtB of E. coli. | | Descriptor: | ACETATE ION, Probable ammonium transporter, SULFATE ION | | Authors: | Zheng, L, Kostrewa, D, Berneche, S, Winkler, F.K, Li, X.-D. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XQF
 
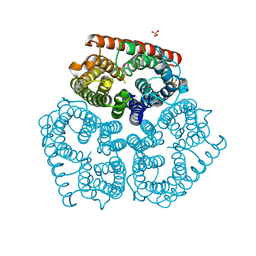 | | The mechanism of ammonia transport based on the crystal structure of AmtB of E. coli. | | Descriptor: | ACETATE ION, Probable ammonium transporter, SULFATE ION | | Authors: | Zheng, L, Kostrewa, D, Berneche, S, Winkler, F.K, Li, X.-D. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3EAD
 
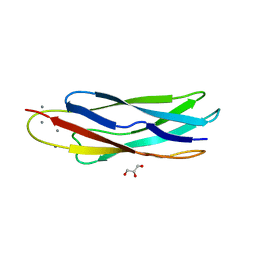 | | Crystal structure of CALX-CBD1 | | Descriptor: | CALCIUM ION, GLYCEROL, Na/Ca exchange protein | | Authors: | Zheng, L, Wang, M. | | Deposit date: | 2008-08-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
3DAE
 
 | |
7W27
 
 | | Crystal structure of BEND3-BEN4-DNA complex | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*CP*GP*TP*GP*GP*GP*TP*C)-3') | | Authors: | Zheng, L, Ren, A. | | Deposit date: | 2021-11-22 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Distinct structural bases for sequence-specific DNA binding by mammalian BEN domain proteins.
Genes Dev., 36, 2022
|
|
7S0Z
 
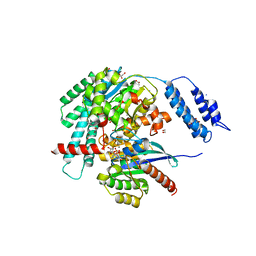 | | Structures of TcdB in complex with R-Ras | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Zheng, L, Rongsheng, J, Peng, C. | | Deposit date: | 2021-08-31 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B.
Sci Adv, 7, 2021
|
|
7S0Y
 
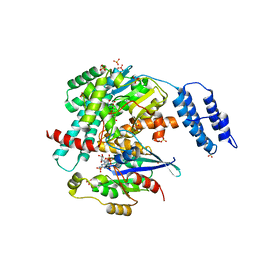 | | Structures of TcdB in complex with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zheng, L, Rongsheng, J, Peng, C. | | Deposit date: | 2021-08-31 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B.
Sci Adv, 7, 2021
|
|
6K61
 
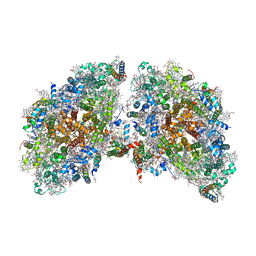 | | Cryo-EM structure of the tetrameric photosystem I from a heterocyst-forming cyanobacterium Anabaena sp. PCC7120 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zheng, L, Li, Y, Li, X, Zhong, Q, Li, N, Zhang, K, Zhang, Y, Chu, H, Ma, C, Li, G, Zhao, J, Gao, N. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria.
Nat.Plants, 5, 2019
|
|
2L5Y
 
 | |
7D72
 
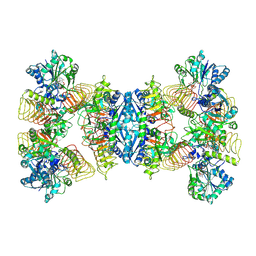 | | Cryo-EM structures of human GMPPA/GMPPB complex bound to GDP-Mannose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D73
 
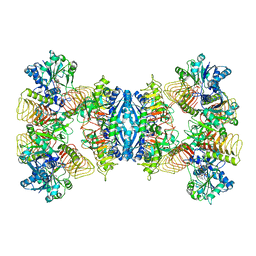 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (State I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D74
 
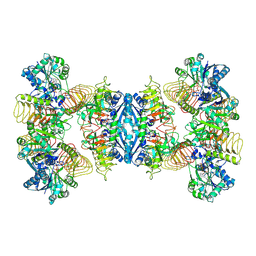 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (state II) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, Mannose-1-phosphate guanyltransferase beta | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-19 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2QZ5
 
 | |
2QZQ
 
 | | Crystal structure of C-terminal of Aida | | Descriptor: | Axin interactor, dorsalization associated protein | | Authors: | Zheng, L.S, Wu, J.W. | | Deposit date: | 2007-08-17 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of C-terminal of Aida
To be Published
|
|
5Y87
 
 | |
5Y85
 
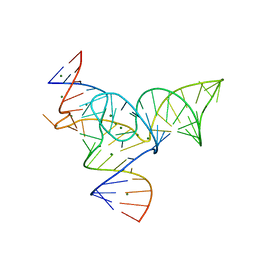 | |
7XFH
 
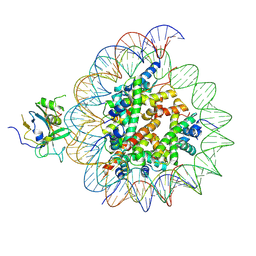 | | Structure of nucleosome-AAG complex (A-30I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFL
 
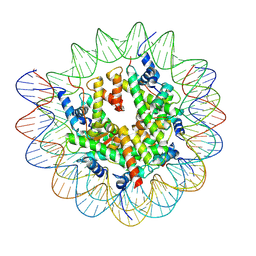 | | Structure of nucleosome-AAG complex (A-53I, free state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFN
 
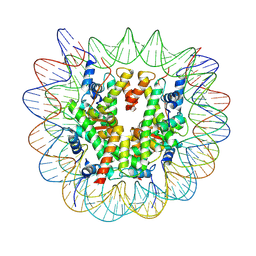 | | Structure of nucleosome-DI complex (-55I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFC
 
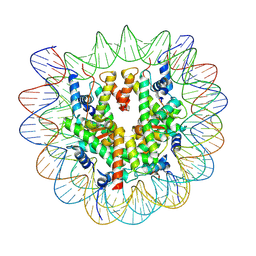 | | Structure of nucleosome-DI complex (-30I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFI
 
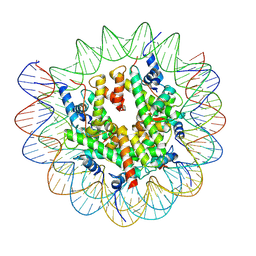 | | Structure of nucleosome-DI complex (-50I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFM
 
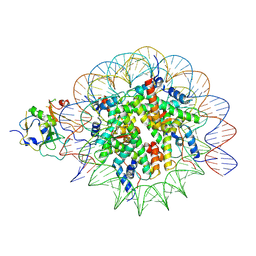 | | Structure of nucleosome-AAG complex (A-53I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
