6U2H
 
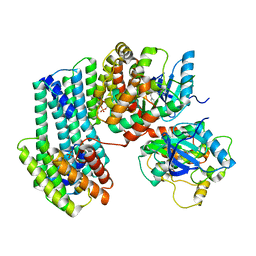 | | BRAF dimer bound to 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6U2G
 
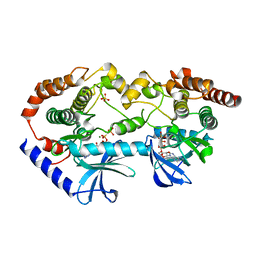 | | BRAF-MEK complex with AMP-PCP bound to BRAF | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liau, N.P.D, Wendorff, T, Hymowitz, S, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6XAG
 
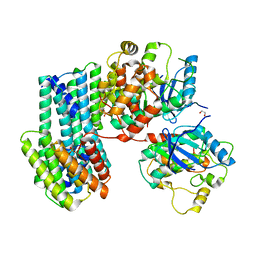 | | Apo BRAF dimer bound to 14-3-3 | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dimerization Induced by C-Terminal 14-3-3 Binding Is Sufficient for BRAF Kinase Activation.
Biochemistry, 59, 2020
|
|
6C7Y
 
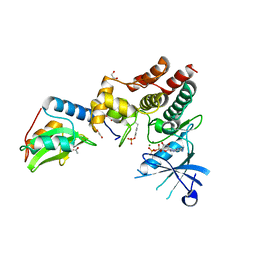 | | Crystal structure of inhibitory protein SOCS1 in complex with JAK1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Liau, N.P.D, Laktyushin, A, Lucet, I.S, Murphy, J.M, Yao, S, Callaghan, K, Nicola, N.A, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | The molecular basis of JAK/STAT inhibition by SOCS1.
Nat Commun, 9, 2018
|
|
7SD1
 
 | |
7SD0
 
 | | Cryo-EM structure of the SHOC2:PP1C:MRAS complex | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liau, N.P.D, Johnson, M.C, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for SHOC2 modulation of RAS signalling.
Nature, 609, 2022
|
|
8G0M
 
 | | Structure of complex between TV6.6 and CD98hc ECD | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | Authors: | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
8DVN
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DVM
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DVL
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.18 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
6C5X
 
 | | Crystal Structure of SOCS1 in complex with ElonginB and ElonginC | | Descriptor: | Elongin-B, Elongin-C, GP130 peptide fragment, ... | | Authors: | Kershaw, N.J, Laktyushin, A, Babon, J.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | The molecular basis of JAK/STAT inhibition by SOCS1.
Nat Commun, 9, 2018
|
|
7SZR
 
 | | NIK bound to inhibitor G02792917 | | Descriptor: | 1-(3-{[(1R,4R,5S)-4-hydroxy-2-methyl-3-oxo-2-azabicyclo[3.1.0]hexan-4-yl]ethynyl}phenyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Liau, N.P.D, Hymowitz, S.G. | | Deposit date: | 2021-11-29 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Filling a nick in NIK: Extending the half-life of a NIK inhibitor through structure-based drug design.
Bioorg.Med.Chem.Lett., 89, 2023
|
|
6XFP
 
 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | Descriptor: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
7SI1
 
 | |
7SHV
 
 | | Crystal structure of BRAF kinase domain bound to GDC0879 | | Descriptor: | 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Kung, J.E, Sudhamsu, J. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Clearing the Path to Rapid High-Quality Protein Purification
To Be Published
|
|
8EXI
 
 | |
8EXJ
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain in complex with a JAK1 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK1 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EXM
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with a JAK3 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK3 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EXN
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with TYK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-receptor tyrosine-protein kinase TYK2 activation loop peptide, PHOSPHATE ION, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8F88
 
 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with monophosphorylated JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EYA
 
 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with a JAK2 activation loop phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EYC
 
 | |
8EYB
 
 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2 activation loop phosphopeptide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EXK
 
 | |
