6V94
 
 | |
6V9J
 
 | |
6V65
 
 | | Crystal structure of KRAS(GMPPNP)-NF1(GRD)-SPRED1 complex | | Descriptor: | FORMIC ACID, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Yan, W, Simanshu, D.K. | | Deposit date: | 2019-12-04 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Structural Insights into the SPRED1-Neurofibromin-KRAS Complex and Disruption of SPRED1-Neurofibromin Interaction by Oncogenic EGFR.
Cell Rep, 32, 2020
|
|
5DHF
 
 | |
5DIS
 
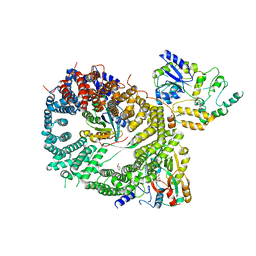 | | Crystal structure of a CRM1-RanGTP-SPN1 export complex bound to a 113 amino acid FG-repeat containing fragment of Nup214 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Monecke, T, Port, S.A, Dickmanns, A, Kehlenbach, R.H, Ficner, R. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export.
Cell Rep, 13, 2015
|
|
6V9M
 
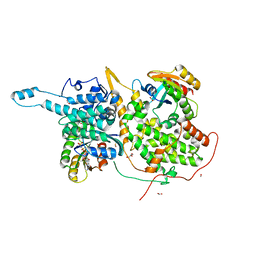 | |
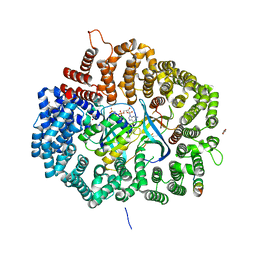
5DH9
 
 | |
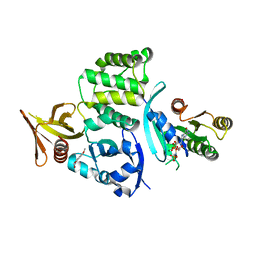
6UZK
 
 | |
6V9F
 
 | |
6V9O
 
 | |
6V9L
 
 | |
5DHA
 
 | |
5DIF
 
 | |
8F7A
 
 | | Cryo-EM structure of Importin-9 bound to RanGTP | | Descriptor: | GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, Importin-9, ... | | Authors: | Bernardes, N.E, Chook, Y.M. | | Deposit date: | 2022-11-18 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Mechanism of RanGTP priming H2A-H2B release from Kap114 in an atypical RanGTP•Kap114•H2A-H2B complex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5DI9
 
 | |
6TKZ
 
 | |
6TGC
 
 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
8HQ6
 
 | | KL2 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HQ3
 
 | | KL1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
6VJJ
 
 | | Crystal Structure of wild-type KRAS4b (GMPPNP-bound) in complex with RAS-binding domain (RBD) of RAF1/CRAF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6W4E
 
 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8HQ4
 
 | | B27 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-fluorophenyl)carbonylamino]-4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Lei, Y, Sun, Q. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | B27 in complex with CRM1-Ran-RanBP1
To Be Published
|
|
8HQ5
 
 | | G6 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-bromophenyl)carbonylamino]-4-[4-(5-chloranyl-2-methyl-phenyl)piperazin-1-yl]benzoic acid, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | F1 in complex with CRM1-Ran-RanBP1
To Be Published
|
|
5DLQ
 
 | | Crystal structure of RanGTP-Exportin 4-eIF5A complex | | Descriptor: | Eukaryotic translation initiation factor 5A-1, Exportin-4, GTP-binding nuclear protein Ran, ... | | Authors: | Aksu, M, Trakhanov, S, Gorlich, D. | | Deposit date: | 2015-09-07 | | Release date: | 2016-06-22 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A.
Nat Commun, 7, 2016
|
|
