3IAR
 
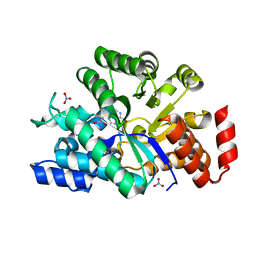 | | The crystal structure of human adenosine deaminase | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, Adenosine deaminase, GLYCEROL, ... | | Authors: | Ugochukwu, E, Zhang, Y, Hapka, E, Yue, W.W, Bray, J.E, Muniz, J, Burgess-Brown, N, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The crystal structure of human adenosine deaminase
To be Published
|
|
7YG1
 
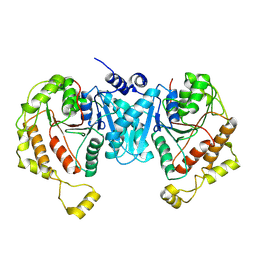 | | Cryo-EM structure of the C-terminal domain of the human sodium-chloride cotransporter | | Descriptor: | Solute carrier family 12 member 3 | | Authors: | Nan, J, Yang, X.M, Shan, Z.Y, Yuan, Y.F, Zhang, Y.Q. | | Deposit date: | 2022-07-09 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structure of the human sodium-chloride cotransporter NCC.
Sci Adv, 8, 2022
|
|
7YG0
 
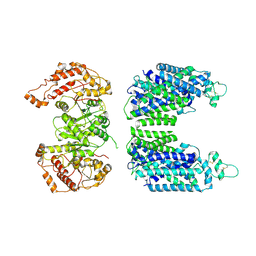 | | Cryo-EM structure of human sodium-chloride cotransporter | | Descriptor: | Solute carrier family 12 member 3 | | Authors: | Nan, J, Yang, X.M, Shan, Z.Y, Yuan, Y.F, Zhang, Y.Q. | | Deposit date: | 2022-07-09 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structure of the human sodium-chloride cotransporter NCC.
Sci Adv, 8, 2022
|
|
7WDL
 
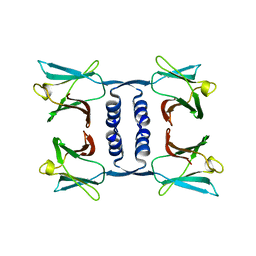 | | Fungal immunomodulatory protein FIP-nha | | Descriptor: | Fungal immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
3IYP
 
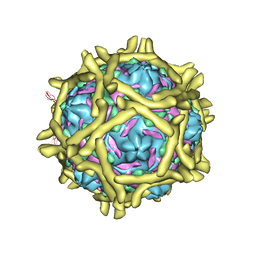 | | The Interaction of Decay-accelerating Factor with Echovirus 7 | | Descriptor: | Capsid protein, Complement decay-accelerating factor, LAURIC ACID, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Harris, K.G, Cifuente, J.O, Bowman, V.D, Chipman, P.R, Lin, F, Medof, D.E, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-04-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Interaction of decay-accelerating factor with echovirus 7.
J.Virol., 84, 2010
|
|
7EPZ
 
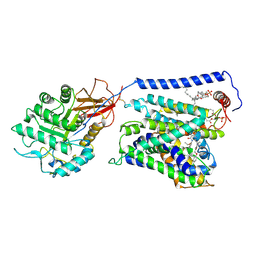 | | Overall structure of Erastin-bound xCT-4F2hc complex | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-[(1S)-1-[4-[2-(4-chloranylphenoxy)ethanoyl]piperazin-1-yl]ethyl]-3-(2-ethoxyphenyl)quinazolin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of erastin-bound xCT-4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis.
Cell Res., 32, 2022
|
|
8IP0
 
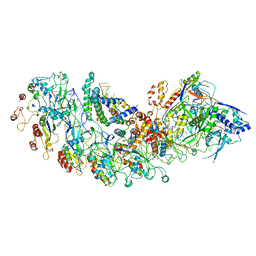 | | Cryo-EM structure of type I-B Cascade bound to a PAM-containing dsDNA target at 3.6 angstrom resolution | | Descriptor: | CRISPR associated protein Cas11b, DNA (41-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and genome editing of type I-B CRISPR-Cas.
Nat Commun, 15, 2024
|
|
3UAZ
 
 | | Crystal structure of Bacillus cereus adenosine phosphorylase D204N mutant complexed with inosine | | Descriptor: | GLYCEROL, INOSINE, Purine nucleoside phosphorylase deoD-type, ... | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UAV
 
 | | Crystal structure of adenosine phosphorylase from Bacillus cereus | | Descriptor: | GLYCEROL, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | Authors: | Dessanti, P, Zhang, Y, Allegrini, S, Tozzi, M.G, Sgarrella, F, Ealick, S.E. | | Deposit date: | 2011-10-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8J3V
 
 | |
8JT3
 
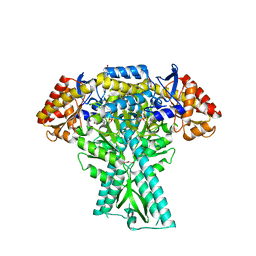 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with amino donor L-Arg | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, ACETATE ION, CrmG, ... | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2023-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Co-crystal structure provides insights on transaminase CrmG recognition amino donor L-Arg.
Biochem.Biophys.Res.Commun., 675, 2023
|
|
7XZO
 
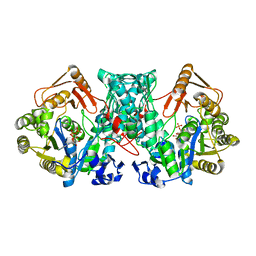 | | Formate-tetrahydrofolate ligase in complex with ATP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | Authors: | Fang, C.L, Zhang, Y. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
7XZN
 
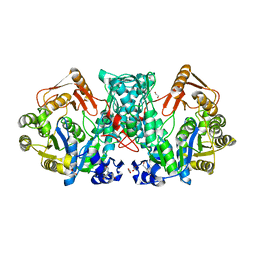 | |
7XZP
 
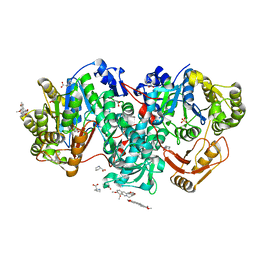 | |
3SOQ
 
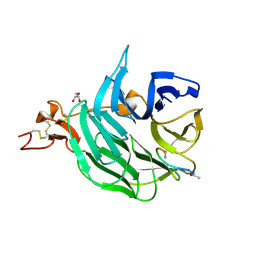 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a DKK1 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dickkopf-related protein 1, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
8H7G
 
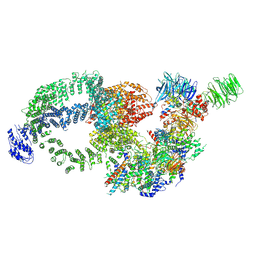 | | Cryo-EM structure of the human SAGA complex | | Descriptor: | Ataxin-7, STAGA complex 65 subunit gamma, Splicing factor 3B subunit 3, ... | | Authors: | Huang, J, Zhang, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of human SAGA transcriptional coactivator complex.
Cell Discov, 8, 2022
|
|
3SOB
 
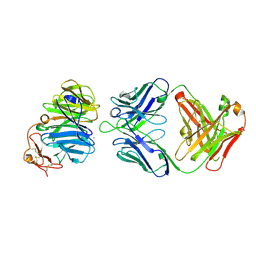 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a FAB | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 6, antibody heavy chain, ... | | Authors: | Wang, W, Bourhis, E, Tam, C, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
7DHS
 
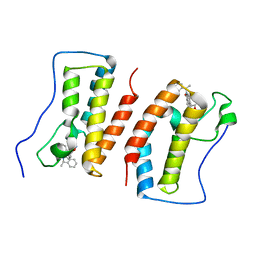 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1R)-1-phenylethyl]benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Wu, T, Xiang, Q, Wang, C, Wu, C, Zhang, C, Zhang, M, Liu, Z, Zhang, Y, Xiao, L, Xu, Y. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Y06014 is a selective BET inhibitor for the treatment of prostate cancer.
Acta Pharmacol.Sin., 42, 2021
|
|
2ITK
 
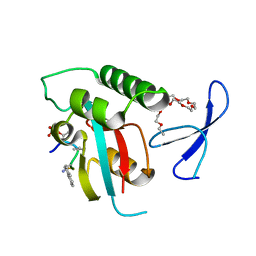 | | human Pin1 bound to D-PEPTIDE | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, D-Peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Noel, J.P, Zhang, Y. | | Deposit date: | 2006-10-19 | | Release date: | 2007-05-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
7YL2
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
3TFP
 
 | | Crystal Structure of Dehydrosqualene Synthase (CrtM) from S. aureus Complexed with BPH-1162 | | Descriptor: | 2-({2-chloro-6-[(2,4-dichlorophenyl)sulfanyl]benzyl}carbamoyl)benzoic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Lin, F.-Y, Liu, Y.-L, Zhang, Y, Oldfield, E. | | Deposit date: | 2011-08-16 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual dehydrosqualene/squalene synthase inhibitors: leads for innate immune system-based therapeutics.
Chemmedchem, 7, 2012
|
|
8KHF
 
 | |
3SOV
 
 | | The structure of a beta propeller domain in complex with peptide S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low-density lipoprotein receptor-related protein 6, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3TFV
 
 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1154 | | Descriptor: | 5-bromo-2-{[3-(octyloxy)benzyl]sulfanyl}benzoic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2011-08-16 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dual dehydrosqualene/squalene synthase inhibitors: leads for innate immune system-based therapeutics.
Chemmedchem, 7, 2012
|
|
2F21
 
 | | human Pin1 Fip mutant | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Bowman, M.E, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-11-15 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
