6LGX
 
 | | Structure of Rabies virus glycoprotein at basic pH | | Descriptor: | Glycoprotein,Glycoprotein,Glycoprotein | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6IGB
 
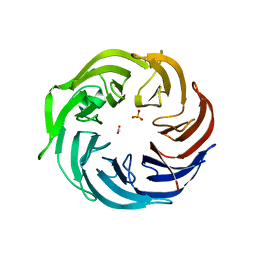 | | the structure of Pseudomonas aeruginosa Periplasmic gluconolactonase, PpgL | | Descriptor: | ACETATE ION, Periplasmic gluconolactonase, PpgL, ... | | Authors: | Song, Y.J, Shen, Y.L, Wang, K.L, Li, T, Zhu, Y.B, Li, C.C, He, L.H, Zhao, N.L, Zhao, C, Yang, J, Huang, Q, Mu, X.Y. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and Functional Insights into PpgL, a Metal-Independent beta-Propeller Gluconolactonase That Contributes toPseudomonas aeruginosaVirulence.
Infect.Immun., 87, 2019
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6TGB
 
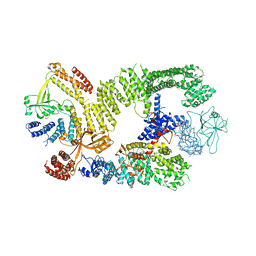 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
3HFH
 
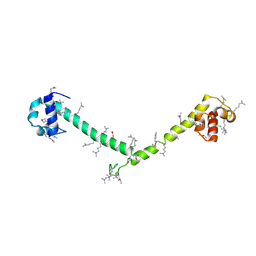 | | Crystal structure of tandem FF domains | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Lu, M, Yang, J, Ren, Z, Subir, S, Bedford, M.T, Jacobson, R.H, McMurray, J.S, Chen, X. | | Deposit date: | 2009-05-11 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of the Three Tandem FF Domains of the Transcription Elongation Regulator CA150.
J.Mol.Biol., 393, 2009
|
|
6TKY
 
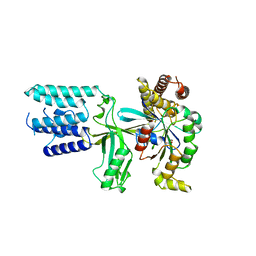 | | Crystal structure of the DHR2 domain of DOCK10 in complex with CDC42 | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 10, GLYCEROL | | Authors: | Barford, D, Fan, D, Cronin, N, Yang, J. | | Deposit date: | 2019-11-29 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for CDC42 and RAC activation by the dual specificity GEF DOCK10
Biorxiv, 2022
|
|
6TM1
 
 | |
6TGC
 
 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
8GOB
 
 | | Crystal Structure of Glycerol Dehydrogenase in the presence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
8GOA
 
 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
5C8Y
 
 | | Crystal structure of T2R-TTL-Plinabulin complex | | Descriptor: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CA1
 
 | | Crystal structure of T2R-TTL-Nocodazole complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
5CA0
 
 | | Crystal structure of T2R-TTL-Lexibulin complex | | Descriptor: | 1-ethyl-3-[2-methoxy-4-(5-methyl-4-{[(1S)-1-(pyridin-3-yl)butyl]amino}pyrimidin-2-yl)phenyl]urea, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-29 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
7D77
 
 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
5CB4
 
 | | Crystal structure of T2R-TTL-Tivantinib complex | | Descriptor: | (3R,4R)-3-(5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1-yl)-4-(1H-indol-3-yl)pyrrolidine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery.
Febs J., 283, 2016
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
8GQW
 
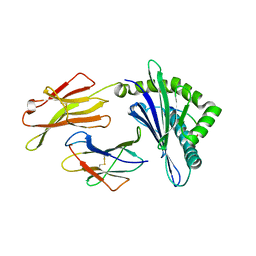 | | The Crystal Structures of a Swine SLA-2*HB01 Molecules Complexed with a CTL epitope from Asia1 serotype of Foot-and-mouth disease virus | | Descriptor: | Hu64, MHC class I antigen, beta 2 microglobulin | | Authors: | Feng, L, Gao, Y.Y, Sun, M.W, Li, Z.B, Zhang, Q, Yang, J, Qiao, C, Jin, H, Feng, H.S, Xian, Y.H, Qi, J.X, Gao, G.F, Liu, W.J, Gao, F.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Parallel Presentation of Two Functional CTL Epitopes Derived from the O and Asia 1 Serotypes of Foot-and-Mouth Disease Virus and Swine SLA-2*HB01: Implications for Universal Vaccine Development.
Cells, 11, 2022
|
|
8GQV
 
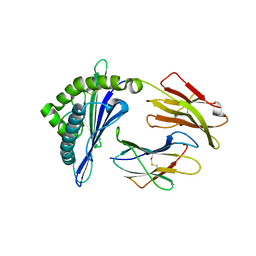 | | The Crystal Structures of a Swine SLA-2*HB01 Molecules Complexed with a CTL epitope from Asia1 serotype of Foot-and-mouth disease virus | | Descriptor: | As64, MHC class I antigen, beta 2 microglobulin | | Authors: | Feng, L, Gao, Y.Y, Sun, M.W, Li, Z.B, Zhang, Q, Yang, J, Qiao, C, Jin, H, Feng, H.S, Xian, Y.H, Qi, J.X, Gao, G.F, Liu, W.J, Gao, F.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Parallel Presentation of Two Functional CTL Epitopes Derived from the O and Asia 1 Serotypes of Foot-and-Mouth Disease Virus and Swine SLA-2*HB01: Implications for Universal Vaccine Development.
Cells, 11, 2022
|
|
6GYP
 
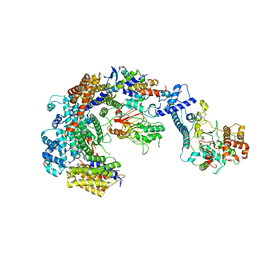 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | Descriptor: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYS
 
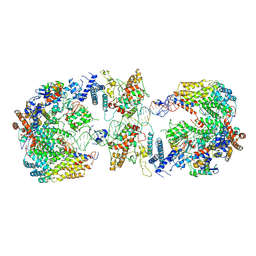 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYU
 
 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-02 | | Release date: | 2018-12-05 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1VYU
 
 | | Beta3 subunit of Voltage-gated Ca2+-channel | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
6IK4
 
 | | A Novel M23 Metalloprotease Pseudoalterin from Deep-sea | | Descriptor: | Elastinolytic metalloprotease, GLYCEROL, ZINC ION | | Authors: | Zhao, H.L, Tang, B.L, Yang, J, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria.
Nat Commun, 11, 2020
|
|
8H24
 
 | | Leucine-rich alpha-2-glycoprotein 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich alpha-2-glycoprotein, SULFATE ION | | Authors: | Won, S.Y, Park, B.S, Lee, D.S, Kim, H.M, Han, A, Yang, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of LRG1 and the functional significance of LRG1 glycan for LPHN2 activation.
Exp.Mol.Med., 55, 2023
|
|
