9C62
 
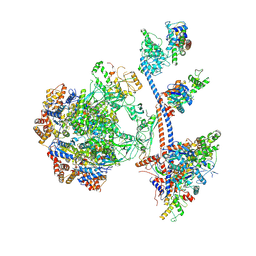 | | P400 subcomplex of the native human TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (5.28 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
1GZ3
 
 | | Molecular mechanism for the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FUMARIC ACID, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Lanks, C.W, Tong, L. | | Deposit date: | 2002-05-14 | | Release date: | 2003-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism for the Regulation of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme by ATP and Fumarate
Structure, 10, 2002
|
|
5FIS
 
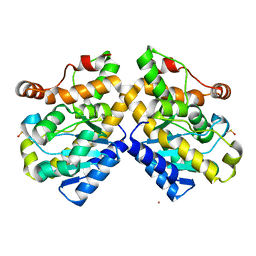 | | Exonuclease domain-containing 1 (Exd1) in the Gd bound conformation | | Descriptor: | EXD1, GADOLINIUM ATOM | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-02 | | Release date: | 2015-12-23 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
9C57
 
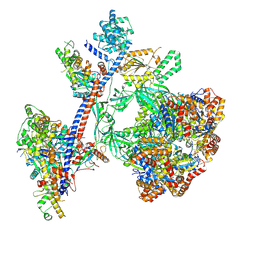 | | Reconstituted P400 Subcomplex of the human TIP60 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-05 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
7RXE
 
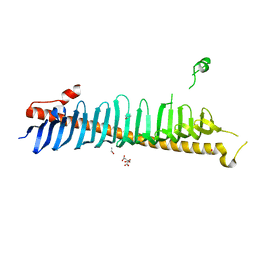 | | Crystal structure of junctophilin-2 | | Descriptor: | CITRATE ANION, ISOPROPYL ALCOHOL, Junctophilin-2 N-terminal fragment | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-22 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RW4
 
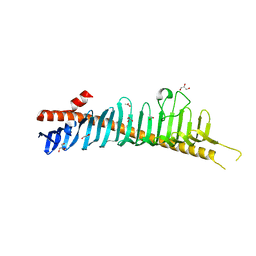 | | Crystal structure of junctophilin-1 | | Descriptor: | ACETATE ION, GLYCEROL, Junctophilin-1 | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RXQ
 
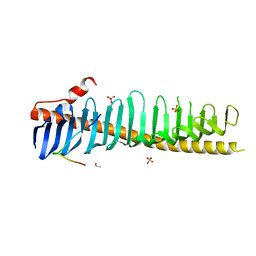 | | Crystal structure of junctophilin-2 in complex with a CaV1.1 peptide | | Descriptor: | ETHANOL, Junctophilin-2 N-terminal fragment, SULFATE ION, ... | | Authors: | Yang, Z, Panwar, P, Van Petegem, F. | | Deposit date: | 2021-08-23 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of the junctophilin/voltage-gated calcium channel interface reveal hot spot for cardiomyopathy mutations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RYV
 
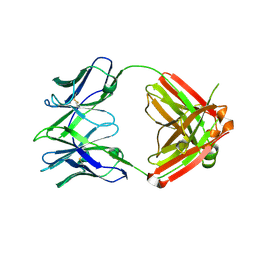 | |
7RYU
 
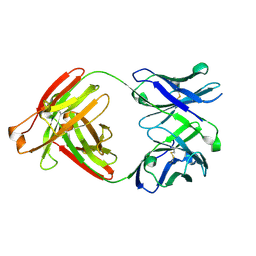 | |
6IJ1
 
 | | Crystal structure of a protein from Actinoplanes | | Descriptor: | ACETATE ION, IMIDAZOLE, Prenylcyclase | | Authors: | Yang, Z.Z, Zhang, L.L, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-10-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Crystal structure of TchmY from Actinoplanes teichomyceticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1C0M
 
 | |
1C1A
 
 | |
1TEH
 
 | |
8D5C
 
 | | anti-HIV-1 gp120-sCD4 complex antibody CG10 Fab in complex with B41-sCD4 | | Descriptor: | CG10 Fab heavy chain, CG10 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | Yang, Z, Bjorkman, P.J, Gershoni, J.M. | | Deposit date: | 2022-06-04 | | Release date: | 2022-11-16 | | Last modified: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Antibody Recognition of CD4-Induced Open HIV-1 Env Trimers.
J.Virol., 96, 2022
|
|
6U0N
 
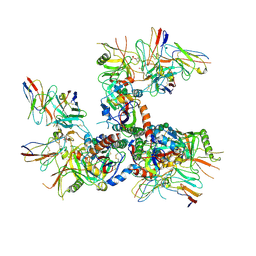 | |
6U0L
 
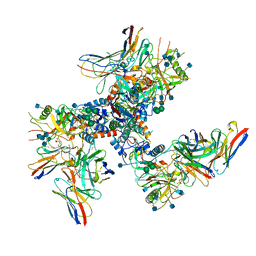 | |
1GQ2
 
 | | Malic Enzyme from Pigeon Liver | | Descriptor: | CHLORIDE ION, MALIC ENZYME, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Zhang, H, Liang, T. | | Deposit date: | 2001-11-19 | | Release date: | 2002-05-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Pigeon Cytosolic Nadp+ -Dependent Malic Enzyme
Protein Sci., 11, 2002
|
|
1GZ4
 
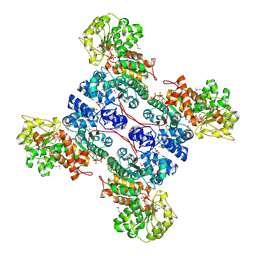 | | molecular mechanism of the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FUMARIC ACID, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Lanks, C.W, Tong, L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Mechanism for the Regulation of Human Mitochondrial Nad(P)(+)-Dependent Malic Enzyme by ATP and Fumarate
Structure, 10, 2002
|
|
1QBZ
 
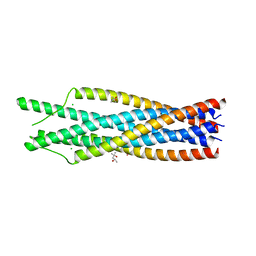 | | THE CRYSTAL STRUCTURE OF THE SIV GP41 ECTODOMAIN AT 1.47 A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CHLORIDE ION, MERCURY (II) ION, ... | | Authors: | Yang, Z.-N, Mueser, T.C, Kaufman, J, Stahl, S.J, Wingfield, P.T, Hyde, C.C. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The crystal structure of the SIV gp41 ectodomain at 1.47 A resolution.
J.Struct.Biol., 126, 1999
|
|
1N86
 
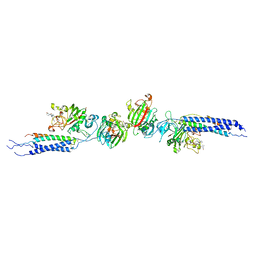 | | Crystal structure of human D-dimer from cross-linked fibrin complexed with GPR and GHRPLDK peptide ligands. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, Fibrin alpha/alpha-E chain, ... | | Authors: | Yang, Z, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-11-19 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of fragment double-D from cross-linked lamprey fibrin reveals isopeptide linkages across an unexpected D-D interface.
Biochemistry, 41, 2002
|
|
1M1J
 
 | | Crystal structure of native chicken fibrinogen with two different bound ligands | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, Z, Kollman, J.M, Pandi, L, Doolittle, R.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Native Chicken Fibrinogen at 2.7 A Resolution
Biochemistry, 40, 2001
|
|
8D54
 
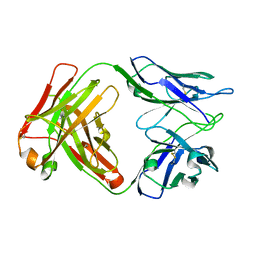 | |
1H2H
 
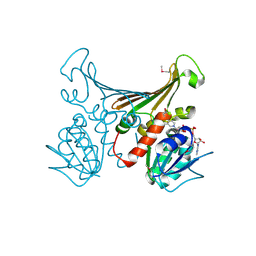 | | Crystal structure of TM1643 | | Descriptor: | HYPOTHETICAL PROTEIN TM1643, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Z, Savchenko, A, Edwards, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-08 | | Release date: | 2002-08-15 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643.
J. Biol. Chem., 278, 2003
|
|
1LWU
 
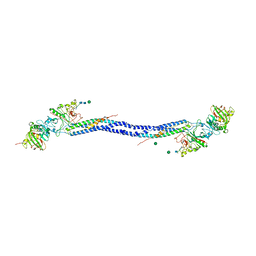 | | Crystal structure of fragment D from lamprey fibrinogen complexed with the peptide Gly-His-Arg-Pro-amide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Spraggon, G, Pandi, L, Everse, S.J, Riley, M, Doolittle, R.F. | | Deposit date: | 2002-06-03 | | Release date: | 2002-08-23 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of fragment D from lamprey fibrinogen complexed with the peptide Gly-His-Arg-Pro-amide.
Biochemistry, 41, 2002
|
|
1DO8
 
 | | CRYSTAL STRUCTURE OF A CLOSED FORM OF HUMAN MITOCHONDRIAL NAD(P)+-DEPENDENT MALIC ENZYME | | Descriptor: | MALIC ENZYME, MANGANESE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Z, Floyd, D.L, Loeber, G, Tong, L. | | Deposit date: | 1999-12-19 | | Release date: | 2000-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a closed form of human malic enzyme and implications for catalytic mechanism.
Nat.Struct.Biol., 7, 2000
|
|
