1FQ4
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN HYDROXYETHYLENE INHIBITOR CP-108,420 AND YEAST ASPARTIC PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(2R)-1-{[(2S,3R,5R)-1-cyclohexyl-3-hydroxy-5-{[2-(morpholin-4-yl)ethyl]carbamoyl}oct-7-yn-2-yl]amino}-3-(methylsulfa nyl)-1-oxopropan-2-yl]-1H-benzimidazole-2-carboxamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1FQ5
 
 | | X-ray structure of a cyclic statine inhibitor PD-129,541 bound to yeast proteinase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(5S,9S,10S,13S)-9-hydroxy-5,10-bis(2-methylpropyl)-4,7,12,16-tetraoxo-3,6,11,17-tetraazabicyclo[17.3.1]tricosa-1(23),19,21-trien-13-yl]-3-(naphthalen-1-yl)-2-(naphthalen-1-ylmethyl)propanamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1FQ6
 
 | | X-RAY STRUCTURE OF GLYCOL INHIBITOR PD-133,450 BOUND TO SACCHAROPEPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1S)-2-{[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]amino}-1-(ethylsulfanyl)-2-oxoethyl]-Nalpha-(morp holin-4-ylsulfonyl)-L-phenylalaninamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-03 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1FQ7
 
 | | X-RAY STRUCTURE OF INHIBITOR CP-72,647 BOUND TO SACCHAROPEPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(tert-butoxycarbonyl)-L-phenylalanyl-N-[(2S,3S,5R)-1-cyclohexyl-3-hydroxy-7-methyl-5-(methylcarbamoyl)octan-2-yl]-L-histidinamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-04 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
1FQ8
 
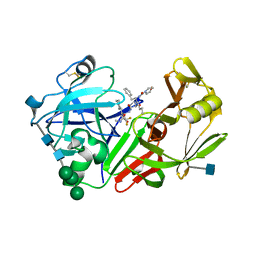 | | X-RAY STRUCTURE OF DIFLUOROSTATINE INHIBITOR CP81,198 BOUND TO SACCHAROPEPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(2S)-1-[[(2S)-1-[[(2S,3R)-1-cyclohexyl-4,4-difluoro-3-hydroxy-5-(methylamino)-5-oxo-pentan-2-yl]amino]-1-oxo-hexan-2 -yl]amino]-1-oxo-3-phenyl-propan-2-yl]morpholine-4-carboxamide, SACCHAROPEPSIN, ... | | Authors: | Cronin, N.B, Badasso, M.O, Tickle, I.J, Dreyer, T, Hoover, D.J, Rosati, R.L, Humblet, C.C, Lunney, E.A, Cooper, J.B. | | Deposit date: | 2000-09-04 | | Release date: | 2000-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structures of five renin inhibitors bound to saccharopepsin: exploration of active-site specificity.
J.Mol.Biol., 303, 2000
|
|
5BPT
 
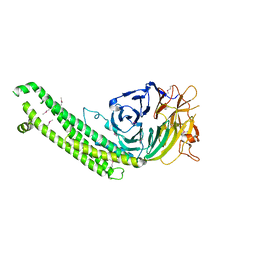 | |
5BPZ
 
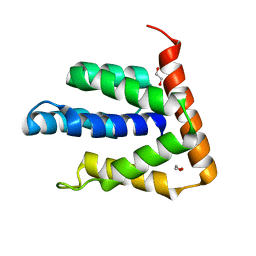 | | Atomic-resolution structures of the APC/C subunits Apc4 and the Apc5 N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Anapc5 protein | | Authors: | Cronin, N, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2015-05-28 | | Release date: | 2015-09-02 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Atomic-Resolution Structures of the APC/C Subunits Apc4 and the Apc5 N-Terminal Domain.
J.Mol.Biol., 427, 2015
|
|
7ZBU
 
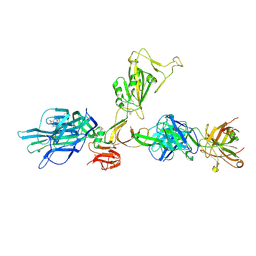 | | CryoEM structure of SARS-CoV-2 spike monomer in complex with neutralising antibody P008_60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, P008_60 antibody, ... | | Authors: | Rosa, A, Pye, V.E, Cronin, N, Cherepanov, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | A neutralizing epitope on the SD1 domain of SARS-CoV-2 spike targeted following infection and vaccination.
Cell Rep, 40, 2022
|
|
3O7W
 
 | | The Crystal Structure of Human Leucine Carboxyl Methyltransferase 1 | | Descriptor: | GLYCEROL, Leucine carboxyl methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsai, M.L, Cronin, N, Djordjevic, S. | | Deposit date: | 2010-08-01 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human leucine carboxyl methyltransferase 1 that regulates protein phosphatase PP2A
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3CL3
 
 | | Crystal Structure of a vFLIP-IKKgamma complex: Insights into viral activation of the IKK signalosome | | Descriptor: | NF-kappa-B essential modulator, ORF K13 | | Authors: | Bagneris, C, Ageichik, A.V, Cronin, N, Boshoff, C, Waksman, G, Barrett, T. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome.
Mol.Cell, 30, 2008
|
|
2JXR
 
 | | STRUCTURE OF YEAST PROTEINASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-(methylamino)-4-oxobutyl]-L-norleucinamide, PROTEINASE A, ... | | Authors: | Aguilar, C.F, Badasso, M, Dreyer, T, Cronin, N.B, Newman, M.P, Cooper, J.B, Hoover, D.J, Wood, S.P, Johnson, M.S, Blundell, T.L. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae.
J.Mol.Biol., 267, 1997
|
|
7OUF
 
 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450 | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[3-(dimethylamino)-3-oxidanylidene-propyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUG
 
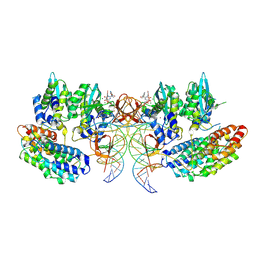 | | STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Integrase, ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUH
 
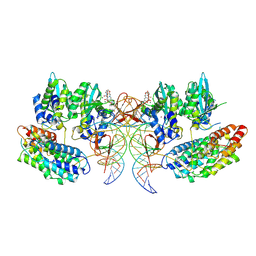 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir | | Descriptor: | Bictegravir, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
5JU5
 
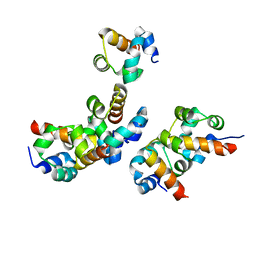 | |
5JTI
 
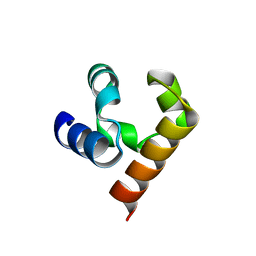 | |
8OJJ
 
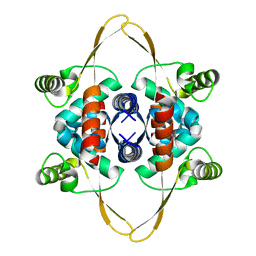 | | Cryo-EM structure of the DnaD-NTD tetramer | | Descriptor: | DNA replication protein DnaD | | Authors: | Winterhalter, C, Pelliciari, S, Cronin, N, Costa, T.R.D, Murray, H, Ilangovan, A. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | The DNA replication initiation protein DnaD recognises a specific strand of the Bacillus subtilis chromosome origin.
Nucleic Acids Res., 51, 2023
|
|
6R6H
 
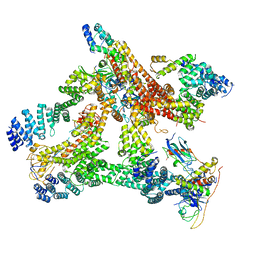 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Morris, E.P, Faull, S.V, Lau, A.M.C, Politis, A, Beuron, F, Cronin, N. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
5JRT
 
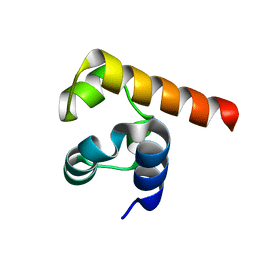 | |
3JQO
 
 | | Crystal structure of the outer membrane complex of a type IV secretion system | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LAURYL DIMETHYLAMINE-N-OXIDE, TraF protein, ... | | Authors: | Chandran, V, Fronzes, R, Duquerroy, S, Cronin, N, Navaza, J, Waksman, G. | | Deposit date: | 2009-09-07 | | Release date: | 2009-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the outer membrane complex of a type IV secretion system
Nature, 462, 2009
|
|
6R7F
 
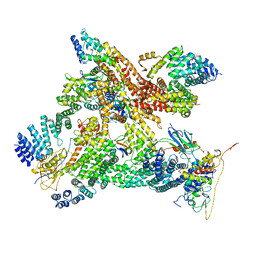 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7H
 
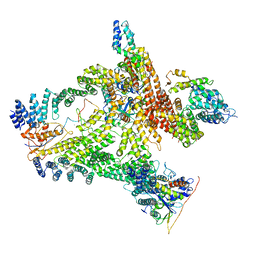 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7N
 
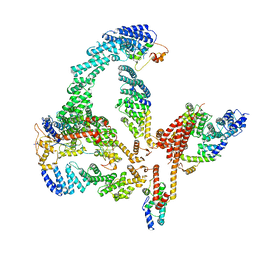 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7I
 
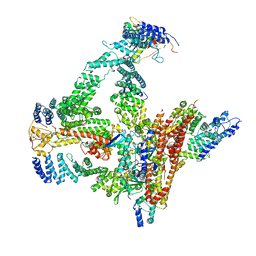 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6TKY
 
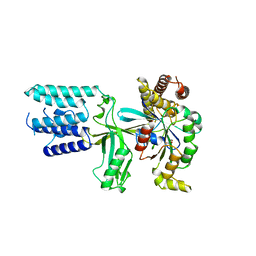 | | Crystal structure of the DHR2 domain of DOCK10 in complex with CDC42 | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 10, GLYCEROL | | Authors: | Barford, D, Fan, D, Cronin, N, Yang, J. | | Deposit date: | 2019-11-29 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for CDC42 and RAC activation by the dual specificity GEF DOCK10
Biorxiv, 2022
|
|
