6UBI
 
 | |
6UDA
 
 | | Cryo-EM structure of CH235UCA bound to Man5-enriched CH505.N279K.G458Y.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH235 UCA heavy chain Fab, CH235 UCA light chain Fab, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancester of CH235 lineage CD4bs broadly neutralizing antibodies.
Plos Pathog., 15, 2019
|
|
5JOF
 
 | |
5U1F
 
 | | Initial contact of HIV-1 Env with CD4: Cryo-EM structure of BG505 DS-SOSIP trimer in complex with CD4 and antibody PGT145 | | Descriptor: | BG505 DS-SOSIP gp120, BG505 SOSIP gp41, PGT145 heavy chain, ... | | Authors: | Acharya, P, Kwong, P.D, Potter, C.S, Carragher, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-02-22 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7LL2
 
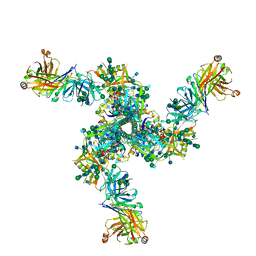 | |
7LL1
 
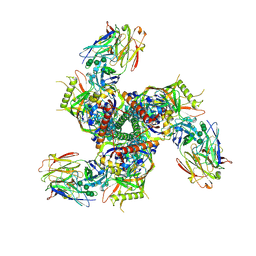 | |
7LQV
 
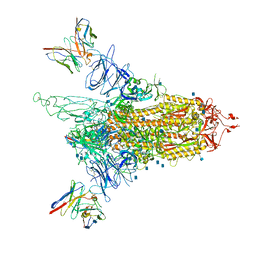 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
7LQW
 
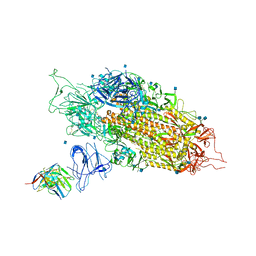 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
7MFG
 
 | | Cryo-EM structure of the VRC310 clinical trial, vaccine-elicited, human antibody 310-030-1D06 Fab in complex with an H1 NC99 HA trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 310-030-1D06 Heavy, 310-030-1D06 Light, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2021-04-09 | | Release date: | 2021-11-03 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | A single residue in influenza virus H2 hemagglutinin enhances the breadth of the B cell response elicited by H2 vaccination.
Nat Med, 28, 2022
|
|
4GW4
 
 | | Crystal structure of 3BNC60 Fab with P61A mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC60 Fab Heavy-chain, 3BNC60 Fab Light-chain | | Authors: | Diskin, R, Fu, B.Z, Bjorkman, P.J. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Somatic Mutations of the Immunoglobulin Framework Are Generally Required for Broad and Potent HIV-1 Neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
6C6Z
 
 | |
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
4I3R
 
 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
7MXP
 
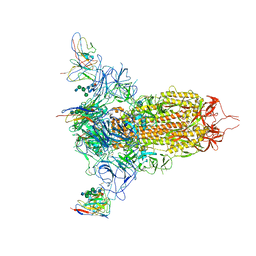 | | Cryo-EM structure of NTD-directed neutralizing antibody LP5 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)][beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Reddem, E.R, Casner, R.G, Shapiro, L. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Antibody screening at reduced pH enables preferential selection of potently neutralizing antibodies targeting SARS-CoV-2.
Aiche J, 67, 2021
|
|
7LG6
 
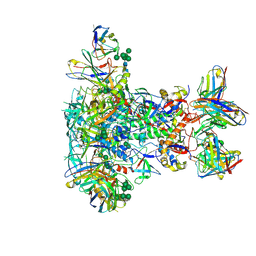 | | BG505 SOSIP.v5.2 in complex with VRC40.01 and RM19R Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Torres, J.L, Wu, N.R, Ward, A.B. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-15 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of glycan276-dependent recognition by HIV-1 broadly neutralizing antibodies.
Cell Rep, 37, 2021
|
|
4I3S
 
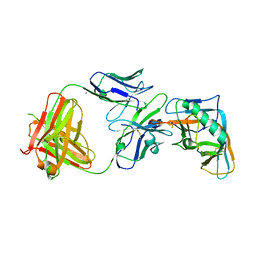 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P21 | | Descriptor: | CALCIUM ION, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
7LLK
 
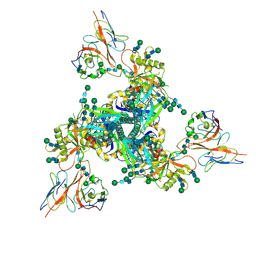 | |
6C6Y
 
 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque in complex with MERS Receptor Binding Domain | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain, SULFATE ION, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
6C6X
 
 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque. | | Descriptor: | JC57-14 Heavy chain, JC57-14 Light chain | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
8RHR
 
 | | E.coli Peptide Deformylase with bound inhibitor BB4 | | Descriptor: | 2-(5-bromo-1H-indol-3-yl)-N-hydroxyacetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Kirschner, H, Stoll, R, Hofmann, E. | | Deposit date: | 2023-12-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Toward More Selective Antibiotic Inhibitors: A Structural View of the Complexed Binding Pocket of E. coli Peptide Deformylase.
J.Med.Chem., 67, 2024
|
|
8TDQ
 
 | | SFX-XFEL structure of CYP121 cocrystallized with substrate cYY | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
8TDP
 
 | | Time-resolved SFX-XFEL crystal structure of CYP121 bound with cYY reacted with peracetic acid for 200 milliseconds | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, HYDROGEN PEROXIDE, Mycocyclosin synthase, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
8TLM
 
 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8U1U
 
 | | Structure of a class A GPCR/agonist complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1,C-C chemokine receptor type 8,EGFP fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8D21
 
 | | Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer | | Descriptor: | 1B06 Heavy Chain, 1B06 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans.
Sci Transl Med, 15, 2023
|
|
