8DS8
 
 | |
6PZV
 
 | |
4GY5
 
 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
7VRF
 
 | | Crystal structure of Oxpecker chromodomain in complex with H3K9me3 | | Descriptor: | H3K9me3, Oxpecker | | Authors: | Huang, Y, Jin, Z, Yu, B. | | Deposit date: | 2021-10-22 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the chromodomain of Oxpecker in complex with histone H3 lysine 9 trimethylation reveal a transposon silencing mechanism by heterodimerization.
Biochem.Biophys.Res.Commun., 652, 2023
|
|
5XPI
 
 | | Structure of UHRF1 TTD in complex with NV01 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, N-[3-(diethylamino)propyl]-2-(12-methyl-9-oxidanylidene-5-thia-1,10,11-triazatricyclo[6.4.0.0^2,6]dodeca-2(6),3,7,11-tetraen-10-yl)ethanamide | | Authors: | Luo, X, Zhao, K. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain
SLAS Discov, 23, 2018
|
|
4LXL
 
 | | Crystal structure of JMJD2B complexed with pyridine-2,4-dicarboxylic acid and H3K9me3 | | Descriptor: | H3 peptide, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | Wang, W.-C, Chu, C.-H, Chen, C.-C. | | Deposit date: | 2013-07-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of JMJD2B complexed with pyridine-2,4-dicarboxylic acid and H3K9me3
To be Published
|
|
4U68
 
 | |
3R93
 
 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
2L75
 
 | |
4YHP
 
 | | Crystal structure of 309M3-B Fab in complex with H3K9me3 peptide | | Descriptor: | Fab Heavy Chain, Fab Light Chain, H3K9me3 peptide | | Authors: | Hattori, T, Dementieva, I.S, Montano, S.P, Koide, S. | | Deposit date: | 2015-02-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Antigen clasping by two antigen-binding sites of an exceptionally specific antibody for histone methylation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4HON
 
 | |
9BAQ
 
 | | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*CP*T)-R(P*(PYO))-D(P*CP*TP*CP*CP*TP*CP*CP*TP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*TP*AP*GP*T)-3'), DNA (cytosine-5-)-methyltransferase, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
3QLA
 
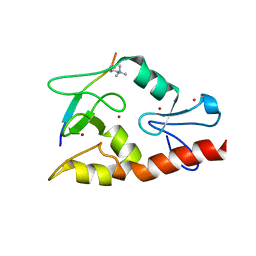 | | Hexagonal complex structure of ATRX ADD bound to H3K9me3 peptide | | Descriptor: | POTASSIUM ION, Transcriptional regulator ATRX, ZINC ION, ... | | Authors: | Xiang, B, Li, H. | | Deposit date: | 2011-02-02 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QL9
 
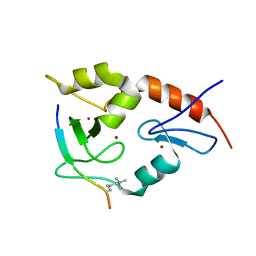 | |
4QUF
 
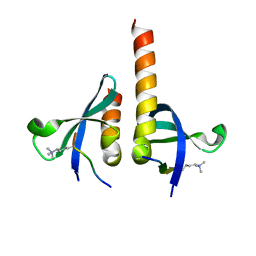 | | crystal structure of chromodomain of Rhino with H3K9me3 | | Descriptor: | H3(1-15)K9me3 peptide, RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
2L12
 
 | | Solution NMR structure of the chromobox protein 7 with H3K9me3 | | Descriptor: | Chromobox homolog 7, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Gutmanas, A, Fares, C, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2L11
 
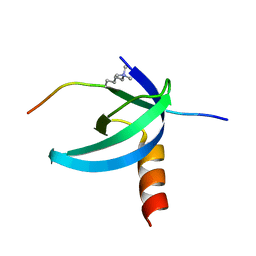 | | Solution NMR structure of the Cbx3 in complex with H3K9me3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Fares, C, Gutmanas, A, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
3GV6
 
 | | Crystal Structure of human chromobox homolog 6 (CBX6) with H3K9 peptide | | Descriptor: | Chromobox protein homolog 6, Histone H3K9me3 peptide | | Authors: | Dong, A, Amaya, M.F, Li, Z, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2Q8C
 
 | | Crystal structure of JMJD2A in ternary complex with an histone H3K9me3 peptide and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, HISTONE 3 PEPTIDE, JmjC domain-containing histone demethylation protein 3A, ... | | Authors: | Couture, J.-F, Collazo, E, Ortiz-Tello, P, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6V41
 
 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
4W5A
 
 | | Complex structure of ATRX ADD bound to H3K9me3S10ph peptide | | Descriptor: | Peptide from Histone H3.3, Transcriptional regulator ATRX, ZINC ION | | Authors: | Zhao, D, Xiang, B, Li, H. | | Deposit date: | 2014-08-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7EBK
 
 | |
6TL1
 
 | | Crystal structure of the TASOR pseudo-PARP domain | | Descriptor: | GLYCEROL, Protein TASOR | | Authors: | Douse, C.H, Timms, R.T, Freund, S.M.V, Modis, Y. | | Deposit date: | 2019-11-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control.
Nat Commun, 11, 2020
|
|
3QLN
 
 | | Crystal structure of ATRX ADD domain in free state | | Descriptor: | Transcriptional regulator ATRX, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
