1VKX
 
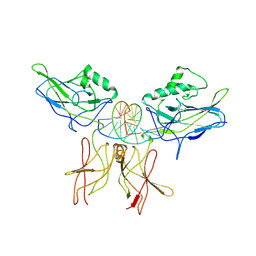 | | CRYSTAL STRUCTURE OF THE NFKB P50/P65 HETERODIMER COMPLEXED TO THE IMMUNOGLOBULIN KB DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*C)-3'), PROTEIN (NF-KAPPA B P50 SUBUNIT), ... | | Authors: | Chen, F, Huang, D.B, Ghosh, G. | | Deposit date: | 1997-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA.
Nature, 391, 1998
|
|
7DKH
 
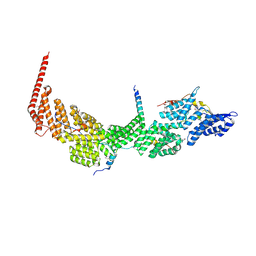 | | Crystal structure of the Ctr9/Paf1/Cdc73/Rtf1 quaternary complex | | Descriptor: | Cell division control protein 73, RNA polymerase II-associated protein 1, RNA polymerase-associated protein CTR9, ... | | Authors: | Chen, F.L, Liu, B.B, Guo, L, Li, D.F, Zhou, H, Long, J.F. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-24 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Core Module of the Yeast Paf1 Complex.
J.Mol.Biol., 434, 2021
|
|
4GU0
 
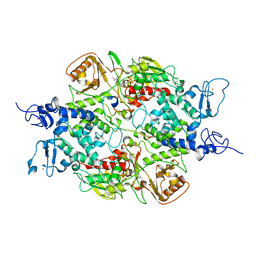 | | Crystal structure of LSD2 with H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Yang, H, Dong, Z, Fang, J, Zhu, T, Gong, W, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b
Cell Res., 23, 2013
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GU1
 
 | | Crystal structure of LSD2 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUU
 
 | | Crystal structure of LSD2-NPAC with tranylcypromine | | Descriptor: | Lysine-specific histone demethylase 1B, Putative oxidoreductase GLYR1, ZINC ION, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4HSU
 
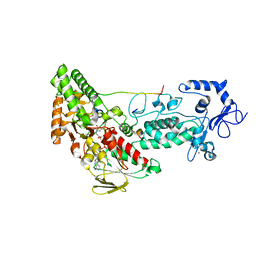 | | Crystal structure of LSD2-NPAC with H3(1-26)in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Xu, Y. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b.
Cell Res., 23, 2013
|
|
6UWN
 
 | |
6UX4
 
 | |
6U6H
 
 | | Calcium-bound MthK open-inactivated state 3 | | Descriptor: | Calcium-gated potassium channel MthK | | Authors: | Chen, F, Crina, N. | | Deposit date: | 2019-08-29 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Ball-and-chain inactivation in a calcium-gated potassium channel.
Nature, 580, 2020
|
|
6UXB
 
 | |
6UXA
 
 | |
6UX7
 
 | |
8OQ7
 
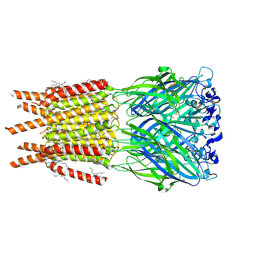 | | CryoEM structure of human rho1 GABAA receptor in complex with inhibitor TPMPA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQA
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA and picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQ6
 
 | | CryoEM structure of human rho1 GABAA receptor apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OP9
 
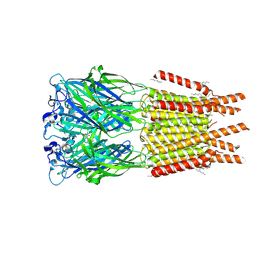 | | CryoEM structure of human rho1 GABAA receptor in complex with GABA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-06 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
8OQ8
 
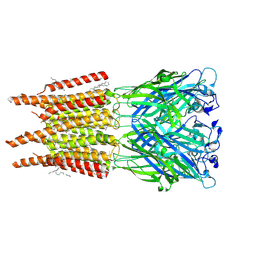 | | CryoEM structure of human rho1 GABAA receptor in complex with pore blocker picrotoxin | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | Authors: | Chen, F, Victor, T, John, C, Rebecca, J.H, Lindahl, E. | | Deposit date: | 2023-04-11 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and dynamics of differential ligand binding in the human rho-type GABA A receptor.
Neuron, 111, 2023
|
|
6R6U
 
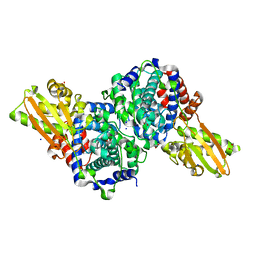 | | Crystal structure of human cis-aconitate decarboxylase | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cis-aconitate decarboxylase, SODIUM ION | | Authors: | Lukat, P, Chen, F, Saile, K, Buessow, K, Pessler, F, Blankenfeldt, W. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Crystal structure ofcis-aconitate decarboxylase reveals the impact of naturally occurring human mutations on itaconate synthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6R6T
 
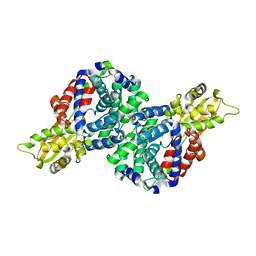 | | Crystal structure of mouse cis-aconitate decarboxylase | | Descriptor: | Cis-aconitate decarboxylase | | Authors: | Lukat, P, Chen, F, Saile, K, Buessow, K, Pessler, F, Blankenfeldt, W. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Crystal structure ofcis-aconitate decarboxylase reveals the impact of naturally occurring human mutations on itaconate synthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1L1V
 
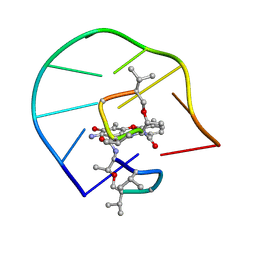 | | UNUSUAL ACTD/DNA_TA COMPLEX STRUCTURE | | Descriptor: | 5'-D(*GP*TP*CP*AP*CP*CP*GP*AP*C)-3', ACTINOMYCIN D | | Authors: | Chou, S.-H, Chin, K.-H, Chen, F.-M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-03-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Looped Out and Perpendicular: Deformation of Watson-Crick Base Pair Associated with Actinomycin D Binding.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1OVF
 
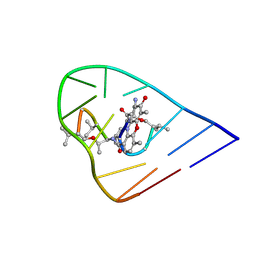 | | NMR Structure of ActD/5'-CCGTTTTGTGG-3' Complex | | Descriptor: | (5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*TP*GP*G)-3'), ACTINOMYCIN D | | Authors: | Chin, K.-H, Chou, S.-H, Chen, F.-M. | | Deposit date: | 2003-03-26 | | Release date: | 2003-05-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Actd-5'-Ccgtt(3)Gtgg-3' Complex: Drug Interaction with Tandem G.T Mismatches and Hairpin Loop Backbone.
Nucleic Acids Res., 31, 2003
|
|
2A7R
 
 | | Crystal structure of human Guanosine Monophosphate reductase 2 (GMPR2) | | Descriptor: | GMP reductase 2, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Li, J, Wei, Z, Zheng, M, Gu, X, Deng, Y, Qiu, R, Chen, F, Ji, C, Gong, W, Xie, Y, Mao, Y. | | Deposit date: | 2005-07-05 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Guanosine Monophosphate Reductase 2 (GMPR2) in Complex with GMP
J.Mol.Biol., 355, 2006
|
|
