1QE1
 
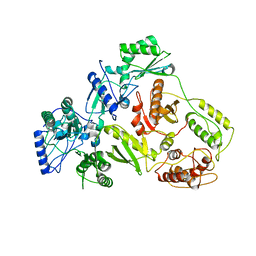 | | CRYSTAL STRUCTURE OF 3TC-RESISTANT M184I MUTANT OF HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | REVERSE TRANSCRIPTASE, SUBUNIT P51, SUBUNIT P66 | | Authors: | Sarafianos, S.G, Das, K, Ding, J, Hughes, S.H, Arnold, E. | | Deposit date: | 1999-07-12 | | Release date: | 1999-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1J5O
 
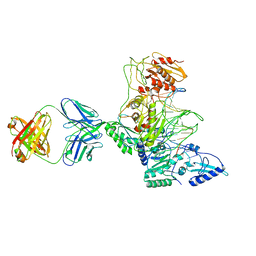 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | Descriptor: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | Authors: | Sarafianos, S.G, Das, K, Arnold, E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1N5Y
 
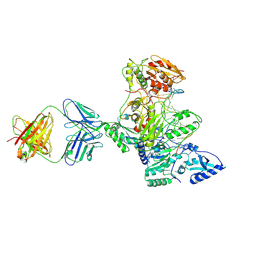 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
1N6Q
 
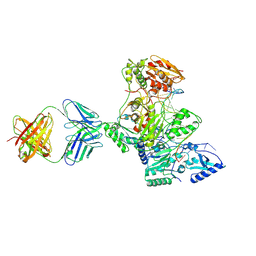 | | HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3', 5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, I, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-11 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA
Embo J., 21, 2002
|
|
1HYS
 
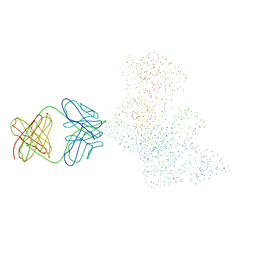 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH A POLYPURINE TRACT RNA:DNA | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*TP*AP*AP*AP*AP*AP*GP*TP*GP*GP*CP*TP*G)-3', 5'-R(*UP*CP*AP*GP*CP*CP*AP*CP*UP*UP*UP*UP*UP*AP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', FAB-28 MONOCLONAL ANTIBODY FRAGMENT HEAVY CHAIN, ... | | Authors: | Sarafianos, S.G, Das, K, Tantillo, C, Clark Jr, A.D, Ding, J, Whitcomb, J, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2001-01-22 | | Release date: | 2001-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA.
EMBO J., 20, 2001
|
|
3NTC
 
 | | Crystal structure of KD-247 Fab, an anti-V3 antibody that inhibits HIV-1 Entry | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fab heavy chain, ... | | Authors: | Sarafianos, S.G, Kirby, K.A. | | Deposit date: | 2010-07-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis of clade-specific HIV-1 neutralization by humanized anti-V3 monoclonal antibody KD-247.
Faseb J., 29, 2015
|
|
3KLF
 
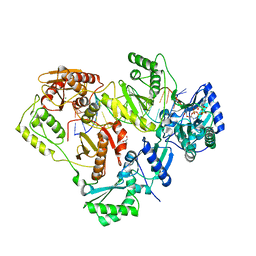 | | Crystal structure of wild-type HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KLE
 
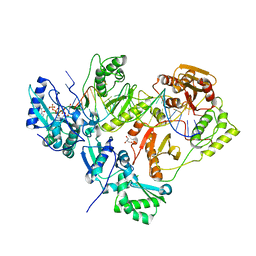 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1ZYR
 
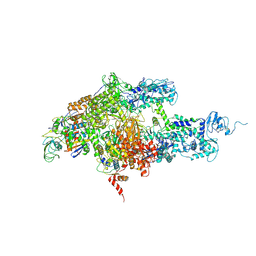 | | Structure of Thermus thermophilus RNA polymerase holoenzyme in complex with the antibiotic streptolydigin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase omega chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-10 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation.
Cell(Cambridge,Mass.), 122, 2005
|
|
4XFX
 
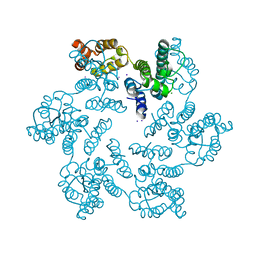 | |
4XFY
 
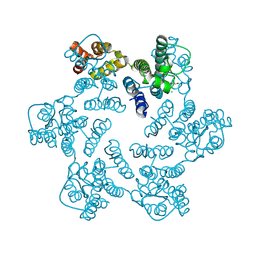 | |
2I5J
 
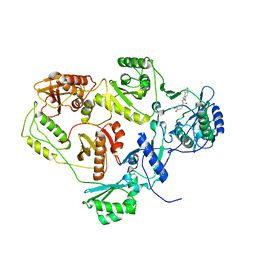 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with DHBNH, an RNASE H inhibitor | | Descriptor: | (E)-3,4-DIHYDROXY-N'-[(2-METHOXYNAPHTHALEN-1-YL)METHYLENE]BENZOHYDRAZIDE, MAGNESIUM ION, Reverse transcriptase/ribonuclease H P51 subunit, ... | | Authors: | Himmel, D.M, Sarafianos, S.G, Knight, J.L, Levy, R.M, Arnold, E. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site.
Acs Chem.Biol., 1, 2006
|
|
6ECL
 
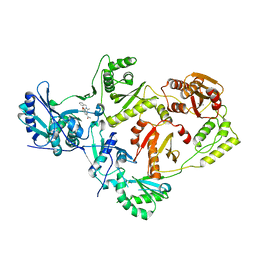 | | Crystal Structure of a 1,2,4-Triazole Allosteric RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 1-[4-methoxy-3-[[5-methyl-4-(phenylmethyl)-1,2,4-triazol-3-yl]sulfanylmethyl]phenyl]ethanone, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Crystal Structure of a 1,2,4-Triazole Allosteric RNase H Inhibitor in Complex with HIV Reverse Transcriptase
To be published
|
|
3KLI
 
 | |
3KLG
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to pre-translocation AZTMP-Terminated DNA (COMPLEX N) | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), Reverse transcriptase/ribonuclease H, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KLH
 
 | | Crystal structure of AZT-Resistant HIV-1 Reverse Transcriptase crosslinked to post-translocation AZTMP-Terminated DNA (COMPLEX P) | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5J2P
 
 | | HIV-1 reverse transcriptase in complex with DNA that has incorporated EFdA-MP at the P-(post-translocation) site and a second EFdA-MP at the N-(pre-translocation) site | | Descriptor: | 2'-deoxy-4'-ethynyl-2-fluoroadenosine 5'-(dihydrogen phosphate), DNA (27-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(6FM)P*(6FM))-3'), ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J2M
 
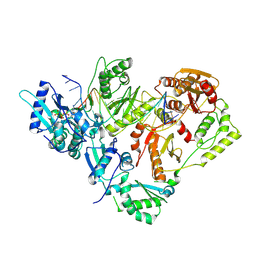 | | HIV-1 reverse transcriptase in complex with DNA and EFdA-triphosphate, a translocation-defective RT inhibitor | | Descriptor: | 2'-deoxy-4'-ethynyl-2-fluoroadenosine 5'-(tetrahydrogen triphosphate), DNA (27-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.432 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4DG1
 
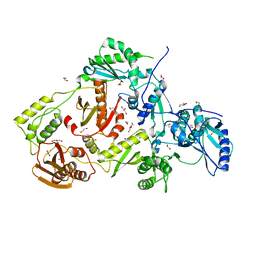 | | Crystal structure of HIV-1 reverse transcriptase (RT) with polymorphism mutation K172A and K173A | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Tu, X, Kirby, K.A, Marchand, B, Sarafianos, S.G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | HIV-1 Reverse Transcriptase (RT) Polymorphism 172K Suppresses the Effect of Clinically Relevant Drug Resistance Mutations to Both Nucleoside and Non-nucleoside RT Inhibitors.
J.Biol.Chem., 287, 2012
|
|
4XFZ
 
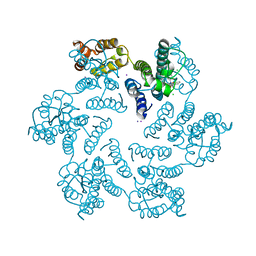 | | Structure of the native full-length HIV-1 capsid protein in complex with PF-3450074 (PF74) | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION, ... | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability.
Science, 349, 2015
|
|
5HBM
 
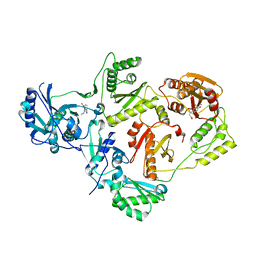 | | Crystal Structure of a Dihydroxycoumarin RNase H Active-Site Inhibitor in Complex with HIV-1 Reverse Transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Crystal Structure of a Dihydroxycoumarin RNase H Active-Site Inhibitor in Complex with HIV-1 Reverse Transcriptase
To Be Published
|
|
5J2N
 
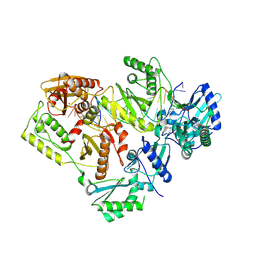 | | HIV-1 reverse transcriptase in complex with DNA that has incorporated EFdA-MP at the P-(post-translocation) site and dTMP at the N-(pre-translocation) site | | Descriptor: | DNA (27-MER), DNA 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', MAGNESIUM ION, ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J2Q
 
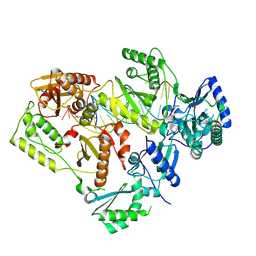 | | HIV-1 reverse transcriptase in complex with DNA that has incorporated a mismatched EFdA-MP at the N-(pre-translocation) site | | Descriptor: | 2'-deoxy-4'-ethynyl-2-fluoroadenosine 5'-(dihydrogen phosphate), DNA (27-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(6FM)P*(6FM))-3'), ... | | Authors: | Salie, Z.L, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structural basis of HIV inhibition by translocation-defective RT inhibitor 4'-ethynyl-2-fluoro-2'-deoxyadenosine (EFdA).
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J1E
 
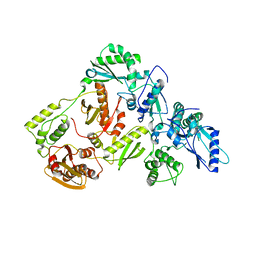 | | Crystal Structure of a Hydroxypyridone Carboxylic Acid Active-Site RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 5-hydroxy-4-oxo-1-[(4'-sulfamoyl[1,1'-biphenyl]-4-yl)methyl]-1,4-dihydropyridine-3-carboxylic acid, HIV-1 REVERSE TRANSCRIPTASE P51 DOMAIN, HIV-1 REVERSE TRANSCRIPTASE P66 DOMAIN, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase H.
J.Med.Chem., 59, 2016
|
|
2CW0
 
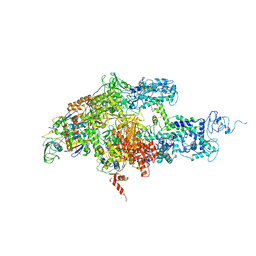 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
