5ER5
 
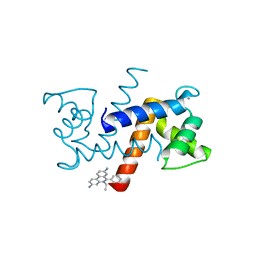 | | Crystal Structure of Calcium-loaded S100B bound to SC1990 | | Descriptor: | CALCIUM ION, ETHIDIUM, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ER4
 
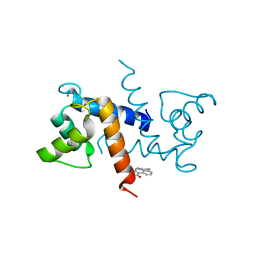 | | Crystal Structure of Calcium-loaded S100B bound to SC0025 | | Descriptor: | 6-methyl-5,6,6~{a},7-tetrahydro-4~{H}-dibenzo[de,g]quinoline-10,11-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5O04
 
 | |
2BGE
 
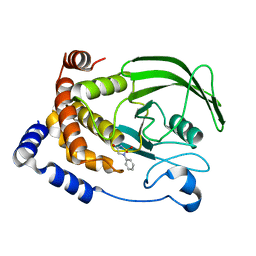 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 1,2,5-THIADIAZOLIDIN-3-ONE-1,1-DIOXIDE, PROTEIN-TYROSINE PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2BGD
 
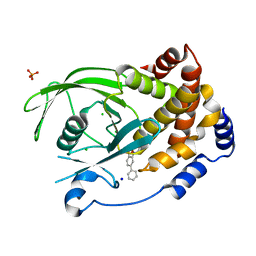 | | Structure-based design of Protein Tyrosine Phosphatase-1B Inhibitors | | Descriptor: | 5-(4-METHOXYBIPHENYL-3-YL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Black, E, Breed, J, Breeze, A.L, Embrey, K, Garcia, R, Gero, T.W, Godfrey, L, Kenny, P.W, Morley, A.D, Minshull, C.A, Pannifer, A.D, Read, J, Rees, A, Russell, D.J, Toader, D, Tucker, J. | | Deposit date: | 2004-12-21 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Protein Tyrosine Phosphatase-1B Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5DKR
 
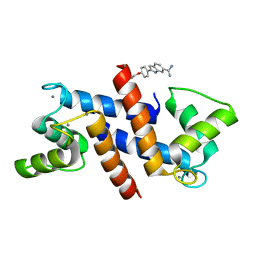 | | Crystal Structure of Calcium-loaded S100B bound to SBi29 | | Descriptor: | 2-[4-(4-carbamimidoylphenoxy)phenyl]-1H-indole-6-carboximidamide, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5DKQ
 
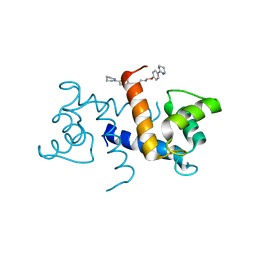 | | Crystal Structure of Calcium-loaded S100B bound to SBi4214 | | Descriptor: | 2,2'-[pentane-1,5-diylbis(oxybenzene-4,1-diyl)]di-1,4,5,6-tetrahydropyrimidine, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5DKN
 
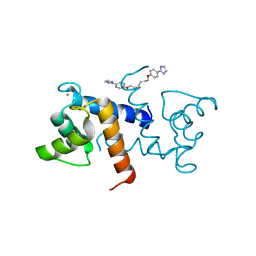 | | Crystal Structure of Calcium-loaded S100B bound to SBi4225 | | Descriptor: | 2,2'-[heptane-1,7-diylbis(oxybenzene-4,1-diyl)]bis(1H-imidazole), CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
7O2W
 
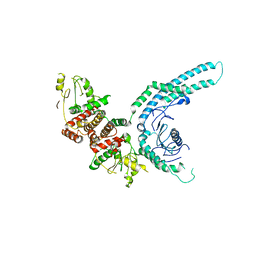 | | Structure of the C9orf72-SMCR8 complex | | Descriptor: | Guanine nucleotide exchange protein SMCR8,Guanine nucleotide exchange protein SMCR8,Maltose/maltodextrin-binding periplasmic protein, Ubiquitin-like protein SMT3,Guanine nucleotide exchange C9orf72 | | Authors: | Noerpel, J, Cavadini, S, Schenk, A.D, Graff-Meyer, A, Chao, J, Bhaskar, V. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture.
Plos Biol., 19, 2021
|
|
7RQ5
 
 | |
4M5W
 
 | | Crystal structure of the USP7/HAUSP catalytic domain | | Descriptor: | BROMIDE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Molland, K.L, Mesecar, A.D, Zhou, Q. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | A 2.2 angstrom resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
5M2H
 
 | | Crystal structure of vancomycin-Zn(II)-citrate complex | | Descriptor: | CITRIC ACID, HEXAETHYLENE GLYCOL, Vancomycin, ... | | Authors: | Zarkan, A, Macklyne, H.-R, Chirgadze, D.Y, Bond, A.D, Hesketh, A.R, Hong, H.-J. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Zn(II) mediates vancomycin polymerization and potentiates its antibiotic activity against resistant bacteria.
Sci Rep, 7, 2017
|
|
1T2M
 
 | | Solution Structure Of The Pdz Domain Of AF-6 | | Descriptor: | AF-6 protein | | Authors: | Zhou, H, Wu, J.H, Xu, Y.Q, Huang, A.D, Shi, Y.Y. | | Deposit date: | 2004-04-22 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of AF-6 PDZ Domain and Its Interaction with the C-terminal Peptides from Neurexin and Bcr
J.Biol.Chem., 280, 2005
|
|
8C3E
 
 | | Engineered mini-protein LCB2 (blocking ligand of SARS-Cov-2 spike protein) | | Descriptor: | Engineered protein LCB2, GLYCEROL | | Authors: | Korban, S.A, Mikhailovskii, O.V, Luzik, D.A, Gurzhiy, V.V, Levkina, A.D, Kharkov, B.B, Skrynnikov, N.R. | | Deposit date: | 2022-12-23 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Using multiple computer-predicted structures as molecular replacement models: application to the antiviral mini-protein LCB2.
Iucrj, 12, 2025
|
|
7JTJ
 
 | |
8OSQ
 
 | |
1S9E
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
8BGC
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with compound 2 (AA-CS-9-003) | | Descriptor: | 5-[(phenylmethyl)amino]pyrimido[4,5-c]quinoline-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Potent and Selective Naphthyridine-Based Chemical Probe for Casein Kinase 2.
Acs Med.Chem.Lett., 14, 2023
|
|
7A6H
 
 | | Cryo-EM structure of human apo RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-08-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1QFG
 
 | | E. COLI FERRIC HYDROXAMATE RECEPTOR (FHUA) | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, DIPHOSPHATE, ... | | Authors: | Ferguson, A.D, Welte, W, Hofmann, E, Lindner, B, Holst, O, Coulton, J.W, Diederichs, K. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
7AE1
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 1 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
9HFY
 
 | | Crystal structure of SARS CoV-2 3CLpro (Mpro) with ALG-097078 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonyl]-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Tauchert, M.J, Maskos, K, McGowan, D.C, Stoycheva, A.D. | | Deposit date: | 2024-11-18 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.282 Å) | | Cite: | Discovery and Preclinical Profile of ALG-097558, a Pan-Coronavirus 3CLpro Inhibitor.
J.Med.Chem., 68, 2025
|
|
9HFX
 
 | | Crystal structure of SARS CoV-2 3CLpro (Mpro) with ALG-097558 | | Descriptor: | (1~{S},2~{S},3~{S},6~{R},7~{R})-~{N}-[(2~{S})-1-azanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-4-[(2~{S})-2-[[2-chloranyl-2,2-bis(fluoranyl)ethanoyl]amino]-3,3-dimethyl-butanoyl]-4-azatricyclo[5.2.1.0^{2,6}]decane-3-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase nsp5 | | Authors: | Tauchert, M.J, Maskos, K, McGowan, D.C, Stoycheva, A.D. | | Deposit date: | 2024-11-18 | | Release date: | 2025-07-16 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery and Preclinical Profile of ALG-097558, a Pan-Coronavirus 3CLpro Inhibitor.
J.Med.Chem., 68, 2025
|
|
