6AYO
 
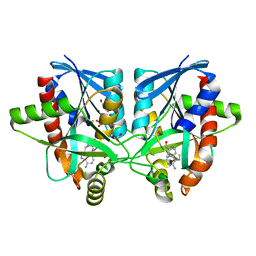 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 5'-deoxy-5'-Propyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-propylpyrrolidin-3-ol, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6EZM
 
 | | Imidazoleglycerol-phosphate dehydratase from Saccharomyces cerevisiae | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2R)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7SA0
 
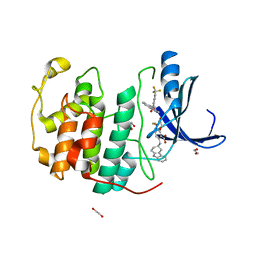 | | Crystal structure of CDK2 liganded with compound EF4195 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(3R)-2,3,4,9-tetrahydro-1H-carbazol-3-yl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-21 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of selective allosteric inhibitors of cyclin-dependent kinase 2 (CDK2)
To Be Published
|
|
5H24
 
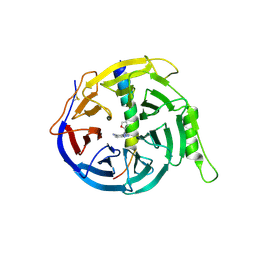 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
7B80
 
 | | DeAMPylation complex of monomeric FICD and AMPylated BiP (state 2) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perera, L.A, Ron, D. | | Deposit date: | 2020-12-12 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of a deAMPylation complex rationalise the switch between antagonistic catalytic activities of FICD.
Nat Commun, 12, 2021
|
|
7YX9
 
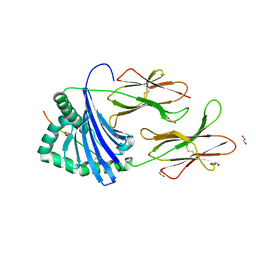 | |
3MY5
 
 | | CDk2/cyclinA in complex with DRB | | Descriptor: | 5,6-dichloro-1-beta-D-ribofuranosyl-1H-benzimidazole, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Baumli, S, Johnson, L.N. | | Deposit date: | 2010-05-09 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Halogen bonds form the basis for selective P-TEFb inhibition by DRB
Chem.Biol., 17, 2010
|
|
6EVQ
 
 | |
8RS7
 
 | |
3NM1
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
7YXB
 
 | | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, CLIP peptide, ... | | Authors: | Roske, Y, Abualrous, E.T, Freund, C. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange.
Nat.Chem.Biol., 19, 2023
|
|
6HB8
 
 | | Crystal structure of OXA-517 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, 2-ETHOXYETHANOL, Beta-lactamase, ... | | Authors: | Raczynska, J.E, Dabos, L, Zavala, A, Retailleau, P, Iorga, B, Jaskolski, M, Naas, T. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Genetic, biochemical and structural characterization of OXA-517, an OXA-48-like extended-spectrum cephalosporins and carbapenems-hydrolyzing beta-lactamase
To Be Published
|
|
6BFE
 
 | | BACE crystal structure with hydroxy pyrrolidine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(1R,2S)-1-[(2R,4R)-4-(cyclohexylmethoxy)pyrrolidin-2-yl]-3-(3,5-difluorophenyl)-1-hydroxypropan-2-yl]acetamide | | Authors: | Timm, D.E. | | Deposit date: | 2017-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Optimization of Hydroxyethylamine Transition State Isosteres as Aspartic Protease Inhibitors by Exploiting Conformational Preferences.
J. Med. Chem., 60, 2017
|
|
7Z98
 
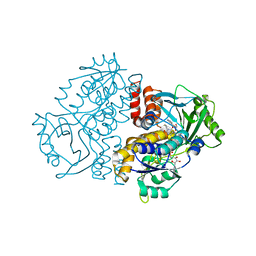 | | Crystal structure of F191M variant Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with methyl phenyl sulfide | | Descriptor: | (methylsulfanyl)benzene, 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6U8A
 
 | |
7YYJ
 
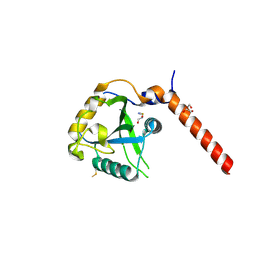 | |
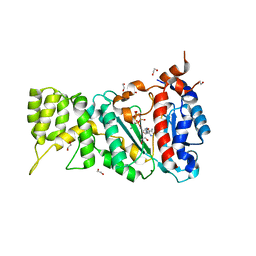
6R5D
 
 | |
6HHR
 
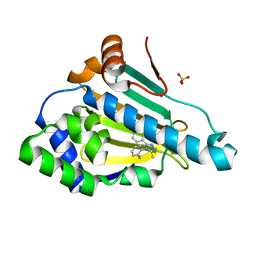 | | Hsp90 in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazole-3-thione | | Descriptor: | 3-[2,4-bis(oxidanyl)phenyl]-4-(2-fluorophenyl)-1~{H}-1,2,4-triazole-5-thione, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehman, M, Eggenweiler, H.-M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Predicting Residence Time and Drug Unbinding Pathway through Scaled Molecular Dynamics.
J.Chem.Inf.Model., 59, 2019
|
|
7YYE
 
 | |
3N0S
 
 | | Crystal structure of BA2930 mutant (H183A) in complex with AcCoA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-06-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
6F3I
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | (3~{R})-3-[4-[[4-(4-ethanoylpiperazin-1-yl)cyclohexyl]amino]pyrrolo[2,1-f][1,2,4]triazin-5-yl]butanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
4O4R
 
 | | Murine Norovirus RdRp in complex with PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, RNA-dependent-RNA-polymerase, SULFATE ION | | Authors: | Croci, R, Tarantino, D, Milani, M, Pezzullo, M, Bolognesi, M, Mastrangelo, E. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PPNDS inhibits murine Norovirus RNA-dependent RNA-polymerase mimicking two RNA stacking bases.
Febs Lett., 588, 2014
|
|
8XBN
 
 | |
8T2H
 
 | | DYRK1A complex with DYR530 | | Descriptor: | (4P)-4-{(3M)-3-[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]-2-methyl-3H-imidazo[4,5-b]pyridin-5-yl}pyridin-2-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL, ... | | Authors: | Montfort, W.R, Basantes, L.E. | | Deposit date: | 2023-06-06 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Functional Characterization of a Potent, Selective, and Metabolically Stable PROTAC of the Protein Kinases DYRK1A and DYRK1B.
J.Med.Chem., 67, 2024
|
|
8X8X
 
 | |
