8JDX
 
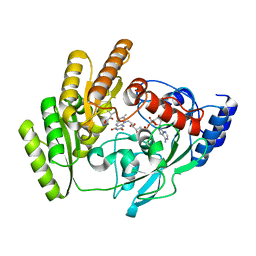 | | Crystal structure of mLDHD in complex with 2-ketoisovaleric acid | | Descriptor: | 3-METHYL-2-OXOBUTANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDG
 
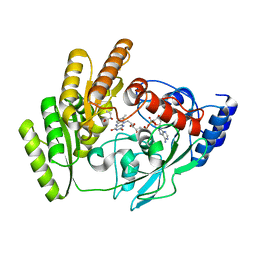 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxybutanoic acid | | Descriptor: | (2R)-2-oxidanylbutanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDR
 
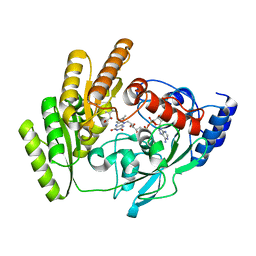 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxy-3-methyl-valeric acid | | Descriptor: | (2R,3S)-3-methyl-2-oxidanyl-pentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDE
 
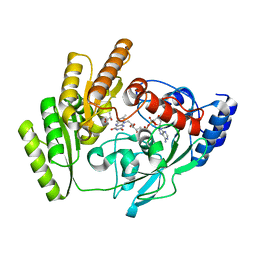 | | Crystal structure of mLDHD in complex with D-lactate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDO
 
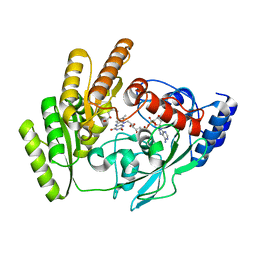 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyhexanoic acid | | Descriptor: | (2R)-2-hydroxyhexanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDU
 
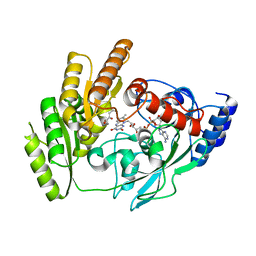 | | Crystal structure of mLDHD in complex with 2-ketovaleric acid | | Descriptor: | 2-oxopentanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDY
 
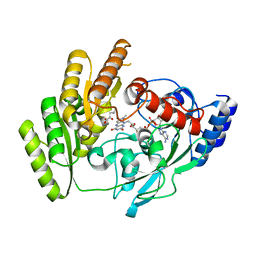 | | Crystal structure of mLDHD in complex with 2-ketoisocaproic acid | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8JDP
 
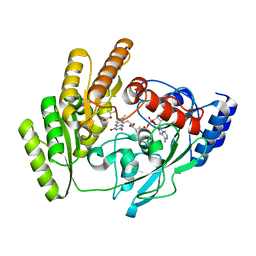 | | Crystal structure of H405A mLDHD in complex with D-2-hydroxyisovaleric acid | | Descriptor: | DEAMINOHYDROXYVALINE, FLAVIN-ADENINE DINUCLEOTIDE, Probable D-lactate dehydrogenase, ... | | Authors: | Jin, S, Chen, X, Yang, J, Ding, J. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis.
Nat Commun, 14, 2023
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVX
 
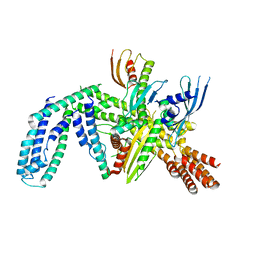 | | Cryo-EM structure of yeast CENP-OPQU+ bound to the CENP-A N-terminus | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CTF19, Inner kinetochore subunit MCM21, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
6OD3
 
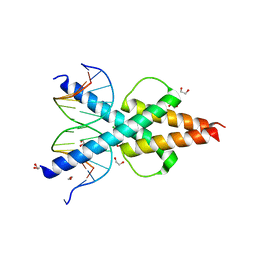 | | Human TCF4 C-terminal bHLH domain in Complex with 13-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*TP*AP*CP*AP*CP*GP*TP*GP*TP*AP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
6P3Q
 
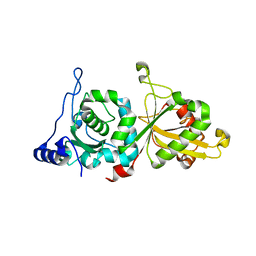 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
6OD5
 
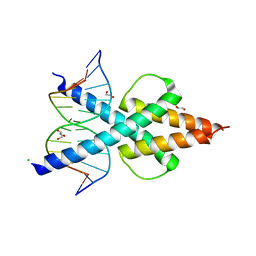 | | Human TCF4 C-terminal bHLH domain in Complex with 12-bp Oligonucleotide Containing E-box Sequence with 5-carboxylcytosines | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*AP*(1CC)P*GP*CP*AP*CP*GP*TP*GP*(1CC)P*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
6OD4
 
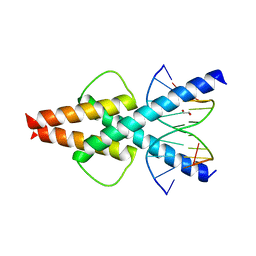 | | Human TCF4 C-terminal bHLH domain in Complex with 11-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*CP*AP*CP*GP*TP*GP*TP*A)-3'), Transcription factor 4 | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
1P93
 
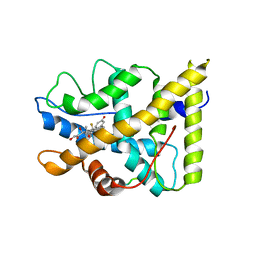 | | CRYSTAL STRUCTURE OF THE AGONIST FORM OF GLUCOCORTICOID RECEPTOR | | Descriptor: | DEXAMETHASONE, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2003-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
7DVM
 
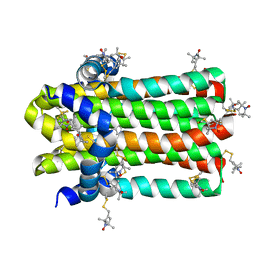 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
6BZ9
 
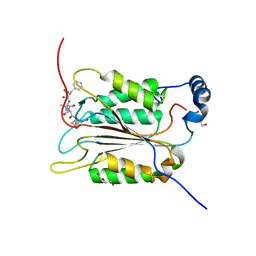 | | Crystal structure of human caspase-1 in complex with Ac-FLTD-CMK | | Descriptor: | Ac-FLTD-CMK, Caspase-1, DI(HYDROXYETHYL)ETHER | | Authors: | Xiao, T.S, Yang, J, Liu, Z, Wang, C, Yang, R. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4AYW
 
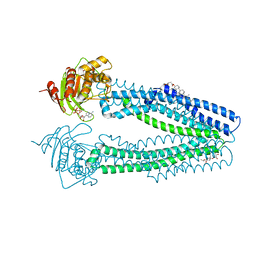 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (PLATE FORM) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3PVB
 
 | | Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Boettcher, A.J, Wu, J, Kim, C, Yang, J, Bruystens, J, Cheung, N, Pennypacker, J.K, Blumenthal, D.A, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2010-12-06 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of (73-244)RIa:C holoenzyme of cAMP-dependent Protein kinase
Structure, 19, 2011
|
|
8J4I
 
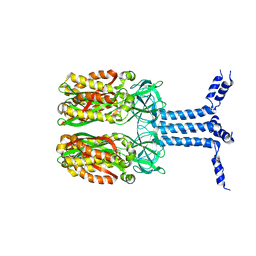 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
6N9N
 
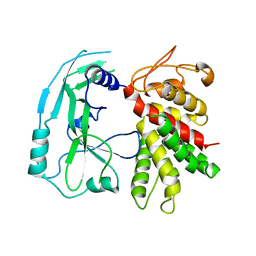 | | Crystal structure of murine GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
6N9O
 
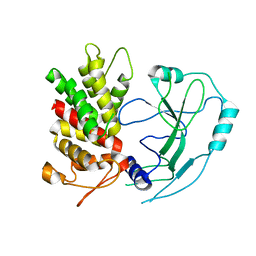 | | Crystal structure of human GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
8X6M
 
 | | Crystal Structure of Glycerol Dehydrogenase in the Presence of NAD+ and Glycerol | | Descriptor: | GLYCEROL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Kang, J.Y, Jin, M, Yang, J, Kim, H, Noh, C, Eom, S.H. | | Deposit date: | 2023-11-21 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the octamerization of glycerol dehydrogenase.
Plos One, 19, 2024
|
|
