7SR9
 
 | | Human alpha-thrombin with 180- and 220- loops replaced with homologous loops from protein C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Di Cera, E, Ruben, E.A, Chen, Z. | | Deposit date: | 2021-11-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The active site region plays a critical role in Na + binding to thrombin.
J.Biol.Chem., 298, 2022
|
|
7TPP
 
 | |
3GIC
 
 | | Structure of thrombin mutant delta(146-149e) in the free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain | | Authors: | Bah, A, Carrell, C.J, Chen, Z, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-03-05 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Stabilization of the E* form turns thrombin into an anticoagulant.
J.Biol.Chem., 284, 2009
|
|
4NZQ
 
 | | Crystal structure of Ca2+-free prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O03
 
 | | Crystal structure of Ca2+ bound prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6V5T
 
 | |
6V64
 
 | | Crystal structure of human thrombin bound to ppack with tryptophans replaced by 5-F-tryptophan | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Ruben, E.A, Chen, Z, Di Cera, E. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 19F NMR reveals the conformational properties of free thrombin and its zymogen precursor prethrombin-2.
J.Biol.Chem., 295, 2020
|
|
8TN9
 
 | | Structural architecture of the acidic region of the B domain of coagulation factor V | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor V | | Authors: | Mohammed, B.M, Basore, K, Summers, B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural architecture of the acidic region of the B domain of coagulation factor V.
J.Thromb.Haemost., 22, 2024
|
|
4RKO
 
 | | Crystal structure of thrombin mutant S195T bound with PPACK | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | Deposit date: | 2014-10-13 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
4RN6
 
 | | Structure of prethrombin-2 mutant s195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Thrombin heavy chain | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Niu, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
4MLF
 
 | | Crystal structure for the complex of thrombin mutant D102N and hirudin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Hirudin variant-1, ... | | Authors: | Vogt, A.D, Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Essential role of conformational selection in ligand binding.
Biophys.Chem., 186C, 2014
|
|
4DT7
 
 | | Crystal structure of thrombin bound to the activation domain QEDQVDPRLIDGKMTRRGDS of protein C | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Pozzi, N, Barranco-Medina, S, Chen, Z, Di Cera, E. | | Deposit date: | 2012-02-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposure of R169 controls protein C activation and autoactivation.
Blood, 120, 2012
|
|
5EDK
 
 | | Crystal structure of prothrombin deletion mutant residues 146-167 ( Form II ). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
8FDG
 
 | |
4RKJ
 
 | | Crystal structure of thrombin mutant S195T (free form) | | Descriptor: | GLYCEROL, POTASSIUM ION, Thrombin heavy chain, ... | | Authors: | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | Deposit date: | 2014-10-13 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
3BEF
 
 | | Crystal structure of thrombin bound to the extracellular fragment of PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Prothrombin | | Authors: | Gandhi, P.S, Bah, A, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-11-17 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural identification of the pathway of long-range communication in an allosteric enzyme.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2PV9
 
 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 4, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PUX
 
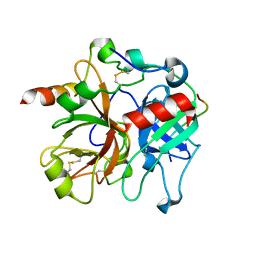 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 3, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2FMJ
 
 | |
2THF
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225F MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, THROMBIN HEAVY CHAIN, ... | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1B7X
 
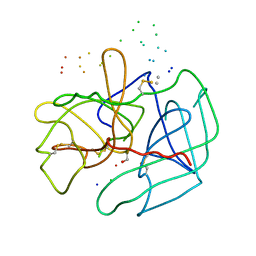 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225I MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | PROTEIN (INHIBITOR), PROTEIN (THROMBIN HEAVY CHAIN), PROTEIN (THROMBIN LIGHT CHAIN) | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-25 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6C2W
 
 | | Crystal structure of human prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
6BJR
 
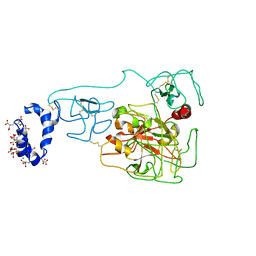 | | Crystal structure of prothrombin mutant S101C/A470C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Prothrombin, ... | | Authors: | Chinnaraj, M, Chen, Z, Pelc, L, Grese, Z, Bystranowska, D, Di Cera, E, Pozzi, N. | | Deposit date: | 2017-11-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structure of prothrombin in the closed form reveals new details on the mechanism of activation.
Sci Rep, 8, 2018
|
|
2GP9
 
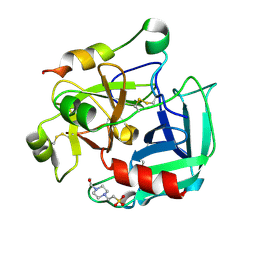 | | Crystal structure of the slow form of thrombin in a self-inhibited conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Prothrombin | | Authors: | Pineda, A, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-04-17 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of thrombin in a self-inhibited conformation.
J.Biol.Chem., 281, 2006
|
|
9C50
 
 | | Replacement of a single residue changes the primary specificity of thrombin | | Descriptor: | FPF, SODIUM ION, Thrombin A-chain, ... | | Authors: | Dei Rossi, A, Deavila, S, Mohammed, B.M, Korolev, S, Di Cera, E. | | Deposit date: | 2024-06-05 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Replacement of a single residue changes the primary specificity of thrombin.
J.Thromb.Haemost., 23, 2025
|
|
