5C8C
 
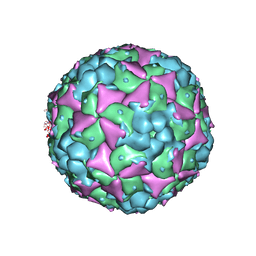 | | Crystal structure of recombinant coxsackievirus A16 capsid | | Descriptor: | CHLORIDE ION, POTASSIUM ION, STEARIC ACID, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
7YUN
 
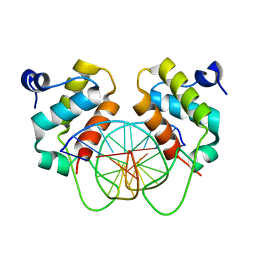 | | Crystal structure of human BEND6 BEN domain in complex with methylated DNA | | Descriptor: | BEN domain-containing protein 6, DNA (5'-D(*CP*TP*CP*TP*CP*GP*(5CM)P*GP*AP*GP*AP*G)-3') | | Authors: | Liu, K, Xiao, Y.Q, Zhang, J, Min, J.R. | | Deposit date: | 2022-08-17 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into DNA recognition by the BEN domain of the transcription factor BANP.
J.Biol.Chem., 299, 2023
|
|
8OVX
 
 | | Cryo-EM structure of yeast CENP-OPQU+ bound to the CENP-A N-terminus | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CTF19, Inner kinetochore subunit MCM21, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
4YLW
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wu, D, Ouyang, P, Lu, W, Huang, J. | | Deposit date: | 2015-03-06 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound
To Be Published
|
|
4KN8
 
 | | Crystal structure of Bs-TpNPPase | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Wang, F, Huang, J, Gong, W, Ji, C. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal Structure of Thermostable p-nitrophenylphosphatase from Bacillus Stearothermophilus (Bs-TpNPPase)
PROTEIN PEPT.LETT., 21, 2014
|
|
2MOX
 
 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
8A5Y
 
 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
8A61
 
 | | S. cerevisiae apo phosphorylated APC/C | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae apo phosphorylated APC/C
To Be Published
|
|
8A3T
 
 | | S. cerevisiae APC/C-Cdh1 complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Vazquez-Fernandez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-09 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex
To Be Published
|
|
5H74
 
 | | Crystal structure of T2R-TTL-14b complex | | Descriptor: | (2~{S},4~{R})-4-[[2-[(1~{R},3~{R})-1-acetyloxy-3-[hexyl-[(2~{S},3~{S})-3-methyl-2-[[(2~{R})-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-4-methyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]-5-(4-fluorophenyl)-2-methyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2016-11-17 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Activity Relationship Studies of Tubulysin Analogues: Anticancer N-Alkyltubulysins with Subpicomolar Activity and the Crystal Structure Binding to Tubulin
To Be Published
|
|
1XMA
 
 | | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833 | | Descriptor: | MERCURY (II) ION, Predicted transcriptional regulator, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chen, L, Lee, D, Habel, J, Nguyen, J, Chang, S.-H, Kataeva, I, Xu, H, Chang, J, Zhao, M, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Praissman, J, Zhang, H, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833
To be published
|
|
6MYV
 
 | | Sialidase26 co-crystallized with DANA-Gc | | Descriptor: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
5CTW
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(butanoylamino)thiophene-3-carboxamide, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Hadfield, A.T, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTX
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPH
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (3E)-3-(pyridin-3-ylmethylidene)-1,3-dihydro-2H-indol-2-one, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-21 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4RSY
 
 | | Crystal structures of the Human leukotriene A4 Hydrolase complex with a potential inhibitor H7 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, ACETIC ACID, IMIDAZOLE, ... | | Authors: | Ouyang, P, Lu, W, Cui, K, Huang, J. | | Deposit date: | 2014-11-12 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of Human Leukotriene A4 Hydrolase in complex with inhibitor H1
To be Published
|
|
4RVB
 
 | | Crystal Structure Analysis of the Human Leukotriene A4 Hydrolase | | Descriptor: | ACETATE ION, ACETIC ACID, GLYCEROL, ... | | Authors: | Ouyang, P, Lu, W, Cui, K, Huang, J. | | Deposit date: | 2014-11-25 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of the Human Leukotriene A4 Hydrolase
To be Published
|
|
8HF0
 
 | | DmDcr-2/R2D2/LoqsPD with 50bp-dsRNA in Dimer state | | Descriptor: | Dicer-2, isoform A, LD06392p, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural mechanism of R2D2 and Loqs-PD synergistic modulation on DmDcr-2 oligomers.
Nat Commun, 14, 2023
|
|
5EZY
 
 | | Crystal structure of T2R-TTL-taccalonolide AJ complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, Y, Yu, Y, Chen, Q, Yang, J. | | Deposit date: | 2015-11-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of microtubule stabilization by taccalonolide AJ
Nat Commun, 8, 2017
|
|
4CSU
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
4U7O
 
 | | Active histidine kinase bound with ATP | | Descriptor: | AMP PHOSPHORAMIDATE, Histidine protein kinase sensor protein | | Authors: | Cai, Y, Hu, X, Sang, J. | | Deposit date: | 2014-07-31 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5CTY
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
