8H7Q
 
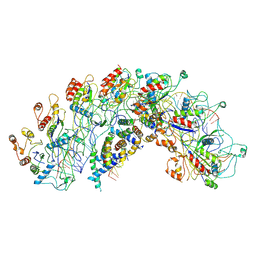 | | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution | | Descriptor: | CRISPR RNA, CRISPR associated protein Cas11b, CRISPR associated protein Cas5, ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC6714 Cascade at 3.8 angstrom resolution
To Be Published
|
|
6AZ3
 
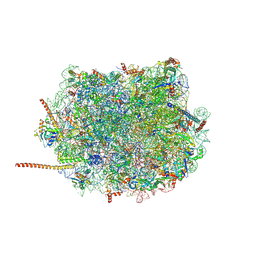 | | Cryo-EM structure of of the large subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | 60S ribosomal protein L10, putative, 60S ribosomal protein L11 (L5, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Nobe, Y, Taoka, M, Matzov, D, Zimmerman, E, Bashan, A, Isobe, T, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
8H67
 
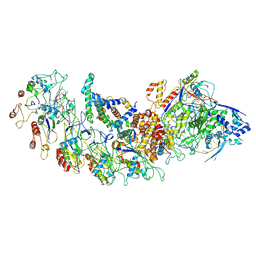 | | type I-B Cascade bound to a PAM-containing dsDNA target at 3.8 angstrom resolution. | | Descriptor: | CRISPR RNA, CRISPR associated protein Cas11b, CRISPR associated protein Cas5, ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2022-10-15 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome editing of type I-B CRISPR-Cas.
Nat Commun, 15, 2024
|
|
6LK8
 
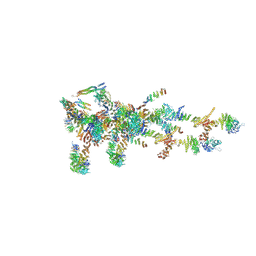 | | Structure of Xenopus laevis Cytoplasmic Ring subunit. | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Shi, Y, Huang, G, Yan, C, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Cell Res., 30, 2020
|
|
5UZ7
 
 | | Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liang, Y.L, Khoshouei, M, Radjainia, M, Zhang, Y, Glukhova, A, Tarrasch, J, Thal, D.M, Furness, S.G.B, Christopoulos, G, Coudrat, T, Danev, R, Baumeister, W, Miller, L.J, Christopoulos, A, Kobilka, B.K, Wootten, D, Skiniotis, G, Sexton, P.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-03 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Phase-plate cryo-EM structure of a class B GPCR-G-protein complex.
Nature, 546, 2017
|
|
8IP0
 
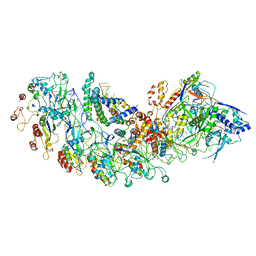 | | Cryo-EM structure of type I-B Cascade bound to a PAM-containing dsDNA target at 3.6 angstrom resolution | | Descriptor: | CRISPR associated protein Cas11b, DNA (41-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Xiao, Y, Lu, M, Yu, C, Zhang, Y. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and genome editing of type I-B CRISPR-Cas.
Nat Commun, 15, 2024
|
|
5XYU
 
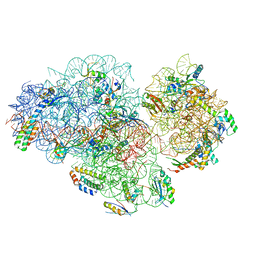 | | Small subunit of Mycobacterium smegmatis ribosome | | Descriptor: | 16S RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Li, Z, Zhang, Y, Zheng, L, Ge, X, Sanyal, S, Gao, N. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis ribosome reveals two unidentified ribosomal proteins close to the functional centers.
Protein Cell, 9, 2018
|
|
6KJ6
 
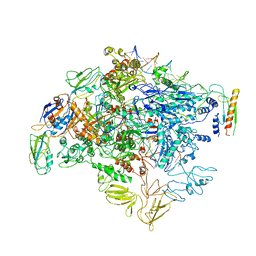 | | cryo-EM structure of Escherichia coli Crl transcription activation complex | | Descriptor: | DNA (51-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Xu, J, Zhang, Y. | | Deposit date: | 2019-07-21 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Crl activates transcription by stabilizing active conformation of the master stress transcription initiation factor.
Elife, 8, 2019
|
|
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
6L94
 
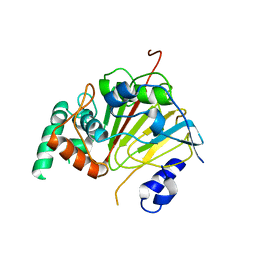 | | The structure of the dioxygenase ABH1 from mouse | | Descriptor: | FE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Xie, W, Wang, C, Li, H, Zhang, Y. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.10012341 Å) | | Cite: | ALKBH1 promotes lung cancer by regulating m6A RNA demethylation.
Biochem Pharmacol, 189, 2021
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
7D77
 
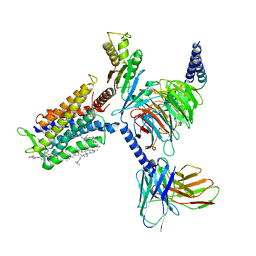 | | Cryo-EM structure of the cortisol-bound adhesion receptor GPR97-Go complex | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6LDI
 
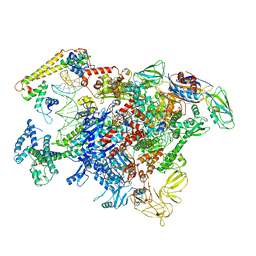 | |
7KOK
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496
to be published
|
|
7EGQ
 
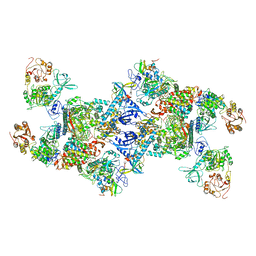 | | Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L.M, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading.
Cell, 184, 2021
|
|
6LH8
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
6LHZ
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
8HYJ
 
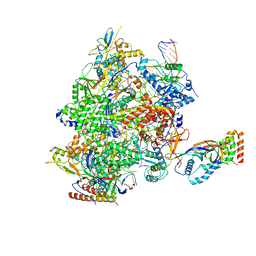 | |
7JN2
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441
to be published
|
|
6AZ1
 
 | | Cryo-EM structure of the small subunit of Leishmania ribosome bound to paromomycin | | Descriptor: | E-site tRNA, LACK1, MAGNESIUM ION, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Rozenberg, H, Matzov, D, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2017-09-09 | | Release date: | 2017-12-06 | | Last modified: | 2019-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Atomic resolution snapshot of Leishmania ribosome inhibition by the aminoglycoside paromomycin.
Nat Commun, 8, 2017
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
