1BOS
 
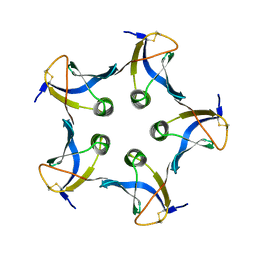 | | SHIGA-LIKE TOXIN COMPLEXED WITH ITS RECEPTOR | | Descriptor: | SHIGA-LIKE TOXIN I B SUBUNIT, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Ling, H, Boodhoo, A, Hazes, B, Cummings, M.D, Armstrong, G.D, Brunton, J.L, Read, R.J. | | Deposit date: | 1998-01-13 | | Release date: | 1999-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3.
Biochemistry, 37, 1998
|
|
1VVJ
 
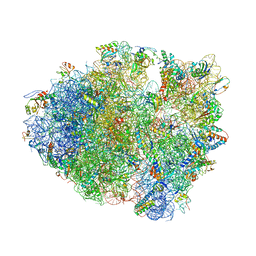 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 bound to Codon CCC-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-05-24 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.44000111574 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1VY4
 
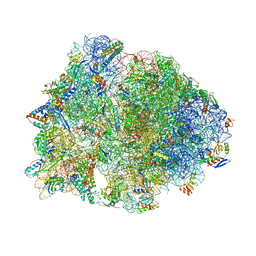 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing acylated tRNA-substrates in the A and P sites. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1VY5
 
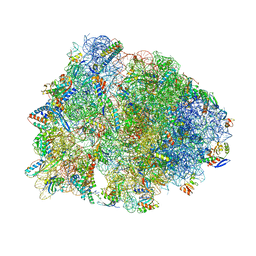 | | Crystal structure of the Thermus thermophilus 70S ribosome in the post-catalysis state of peptide bond formation containing dipeptydil-tRNA in the A site and deacylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1VY6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing short substrate-mimic Cytidine-Puromycin in the A site and acylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1VY7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing short substrate-mimic Cytidine-Cytidine-Puromycin in the A site and acylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2BTJ
 
 | | Fluorescent Protein EosFP - red form | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Photo-Induced Protein Cleavage and Green-to-Red Conversion of Fluorescent Protein Eosfp.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2N6Z
 
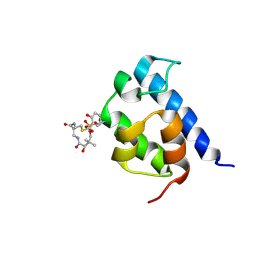 | | Solution structure of the salicylate-loaded ArCP from yersiniabactin synthetase | | Descriptor: | HMWP2 nonribosomal peptide synthetase, S-{2-[(N-{(2R)-2-hydroxy-3,3-dimethyl-4-[(trihydroxy-lambda~5~-phosphanyl)oxy]butanoyl}-beta-alanyl)amino]ethyl} 2-hydroxybenzene-1-carbothioate | | Authors: | Goodrich, A.C, Harden, B.J, Frueh, D.P. | | Deposit date: | 2015-08-31 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Nonribosomal Peptide Synthetase Carrier Protein Loaded with Its Substrate Reveals Transient, Well-Defined Contacts.
J.Am.Chem.Soc., 137, 2015
|
|
2VVJ
 
 | | IrisFP fluorescent protein in its red form, cis conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3BCR
 
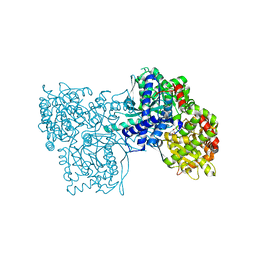 | | Glycogen Phosphorylase b in complex with AZT | | Descriptor: | 3'-azido-3'-deoxythymidine, Glycogen phosphorylase, muscle form | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
3J3Q
 
 | |
3J3Y
 
 | |
3J6B
 
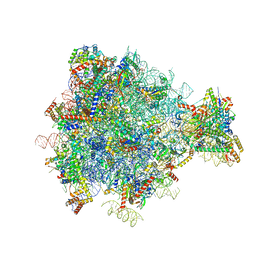 | | Structure of the yeast mitochondrial large ribosomal subunit | | Descriptor: | 21S ribosomal RNA, 54S ribosomal protein IMG1, mitochondrial, ... | | Authors: | Amunts, A, Brown, A, Bai, X.C, Llacer, J.L, Hussain, T, Emsley, P, Long, F, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the yeast mitochondrial large ribosomal subunit.
Science, 343, 2014
|
|
3J6X
 
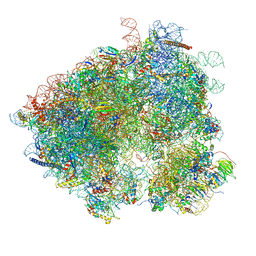 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 5 degree rotation (Class II) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J6Y
 
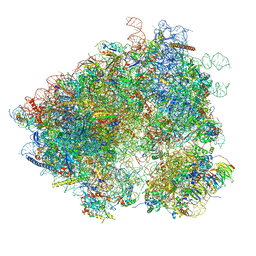 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 2 degree rotation (Class I) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J77
 
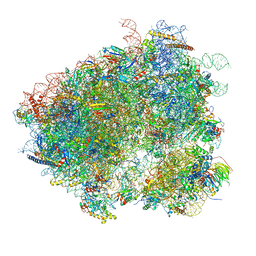 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class II - rotated ribosome with 1 tRNA) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
3J78
 
 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class I - non-rotated ribosome with 2 tRNAs) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
3J79
 
 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, large subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
3J7O
 
 | | Structure of the mammalian 60S ribosomal subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J7P
 
 | | Structure of the 80S mammalian ribosome bound to eEF2 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J7Q
 
 | | Structure of the idle mammalian ribosome-Sec61 complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J7R
 
 | | Structure of the translating mammalian ribosome-Sec61 complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
3J8H
 
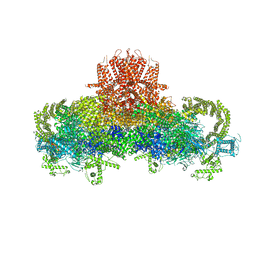 | | Structure of the rabbit ryanodine receptor RyR1 in complex with FKBP12 at 3.8 Angstrom resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Yan, Z, Bai, X, Yan, C, Wu, J, Scheres, S.H.W, Shi, Y, Yan, N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution.
Nature, 517, 2015
|
|
3J92
 
 | | Structure and assembly pathway of the ribosome quality control complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Shao, S, Brown, A, Santhanam, B, Hegde, R.S. | | Deposit date: | 2014-12-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Assembly Pathway of the Ribosome Quality Control Complex.
Mol.Cell, 57, 2015
|
|
3J9G
 
 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | Descriptor: | VipA, VipB | | Authors: | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|







