1E67
 
 | | Zn-Azurin from Pseudomonas aeruginosa | | Descriptor: | AZURIN, NITRATE ION, ZINC ION | | Authors: | Nar, H, Messerschmidt, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-08-16 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization and Crystal Structure of Zinc Azurin, a by-Product of Heterologous Expression in Escherichia Coli of Pseudomonas Aeruginosa Copper Azurin
Eur.J.Biochem., 205, 1992
|
|
8P01
 
 | | Crystal structure of human STING ectodomain in complex with BI 7446, a potent cyclic dinucleotide STING agonist with broad-spectrum variant activity for the treatment of cancer | | Descriptor: | 3-[(1~{R},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9-fluoranyl-18-oxidanyl-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-6~{H}-imidazo[4,5-d]pyridazin-7-one, Stimulator of interferon genes protein | | Authors: | Nar, H. | | Deposit date: | 2023-05-09 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Discovery of BI 7446: A Potent Cyclic Dinucleotide STING Agonist with Broad-Spectrum Variant Activity for the Treatment of Cancer.
J.Med.Chem., 66, 2023
|
|
1E65
 
 | |
1E5Z
 
 | |
1E5Y
 
 | |
1G36
 
 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | 4-{[1-METHYL-5-(2-METHYL-BENZOIMIDAZOL-1-YLMETHYL)-1H-BENZOIMIDAZOL-2-YLMETHYL]-AMINO}-BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Nar, H. | | Deposit date: | 2000-10-23 | | Release date: | 2001-10-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
1G2M
 
 | | FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 4-{[1-METHYL-5-(2-METHYL-BENZOIMIDAZOL-1-YLMETHYL)-1H-BENZOIMIDAZOL-2-YLMETHYL]-AMINO}-BENZAMIDINE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Nar, H. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
1G32
 
 | | THROMBIN INHIBITOR COMPLEX | | Descriptor: | 4-{[1-METHYL-5-(2-METHYL-BENZOIMIDAZOL-1-YLMETHYL)-1H-BENZOIMIDAZOL-2-YLMETHYL]-AMINO}-BENZAMIDINE, HIRUDIN IIB, PROTHROMBIN | | Authors: | Nar, H. | | Deposit date: | 2000-10-23 | | Release date: | 2001-10-23 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
1G2L
 
 | | FACTOR XA INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, [(1-{2[(4-CARBAMIMIDOYL-PHENYLAMINO)-METHYL]-1-METHYL-1H-BENZOIMIDAZOL-5-YL}-CYCLOPROPYL)-PYRIDIN-2-YL-METHYLENEAMINOOXY]-ACETIC ACID ETHYL ESTER | | Authors: | Nar, H. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
1G30
 
 | | THROMBIN INHIBITOR COMPLEX | | Descriptor: | HIRUDIN IIB, PROTHROMBIN, [(1-{2[(4-CARBAMIMIDOYL-PHENYLAMINO)-METHYL]-1-METHYL-1H-BENZOIMIDAZOL-5-YL}-CYCLOPROPYL)-PYRIDIN-2-YL-METHYLENEAMINOOXY]-ACETIC ACID ETHYL ESTER | | Authors: | Nar, H. | | Deposit date: | 2000-10-23 | | Release date: | 2001-10-23 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
5LLS
 
 | | Porcine dipeptidyl peptidase IV in complex with 8-(3-aminopiperidin-1-yl)-7-[(2-bromophenyl)methyl]-1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3~{R})-3-azanylpiperidin-1-yl]-7-[(2-bromophenyl)methyl]-1,3-dimethyl-purine-2,6-dione, ... | | Authors: | Nar, H, Blaesse, M. | | Deposit date: | 2016-07-28 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Comparative Analysis of Binding Kinetics and Thermodynamics of Dipeptidyl Peptidase-4 Inhibitors and Their Relationship to Structure.
J.Med.Chem., 59, 2016
|
|
1JIZ
 
 | | Crystal Structure Analysis of human Macrophage Elastase MMP-12 | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, ZINC ION, ... | | Authors: | Nar, H, Werle, K, Bauer, M.M.T, Dollinger, H, Jung, B. | | Deposit date: | 2001-07-03 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human macrophage elastase (MMP-12) in complex with a hydroxamic acid inhibitor.
J.Mol.Biol., 312, 2001
|
|
1KTS
 
 | | Thrombin Inhibitor Complex | | Descriptor: | 3-({2-[(4-CARBAMIMIDOYL-PHENYLAMINO)-METHYL]-3-METHYL-3H-BENZOIMIDAZOLE-5-CARBONYL}-PYRIDIN-2-YL-AMINO)-PROPIONIC ACID ETHYL ESTER, hirudin IIB, thrombin | | Authors: | Nar, H. | | Deposit date: | 2002-01-17 | | Release date: | 2002-02-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of novel potent nonpeptide thrombin inhibitors.
J.Med.Chem., 45, 2002
|
|
1KTT
 
 | | Thrombin inhibitor complex | | Descriptor: | 4-(5-BENZENESULFONYLAMINO-1-METHYL-1H-BENZOIMIDAZOL-2-YLMETHYL)-BENZAMIDINE, hirudin IIB, thrombin | | Authors: | Nar, H. | | Deposit date: | 2002-01-17 | | Release date: | 2002-02-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of novel potent nonpeptide thrombin inhibitors.
J.Med.Chem., 45, 2002
|
|
1GTP
 
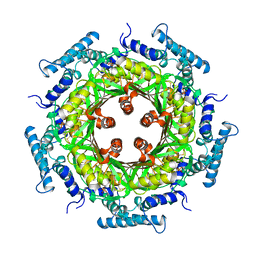 | | GTP CYCLOHYDROLASE I | | Descriptor: | GTP CYCLOHYDROLASE I, SULFATE ION | | Authors: | Nar, H, Huber, R, Meining, W, Bacher, A. | | Deposit date: | 1995-09-16 | | Release date: | 1996-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atomic structure of GTP cyclohydrolase I.
Structure, 3, 1995
|
|
1OYQ
 
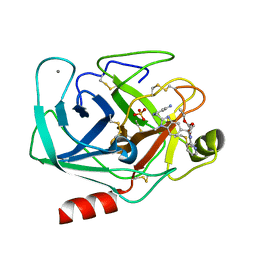 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, SULFATE ION, Trypsin, ... | | Authors: | Nar, H. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhibition Promiscuity of Dual Specific Thrombin and Factor Xa Blood Coagulation Inhibitors
Structure, 9, 2001
|
|
1DB2
 
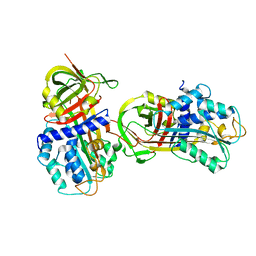 | | CRYSTAL STRUCTURE OF NATIVE PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Nar, H, Bauer, M, Stassen, J.M, Lang, D, Gils, A, Declerck, P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation.
J.Mol.Biol., 297, 2000
|
|
6TL9
 
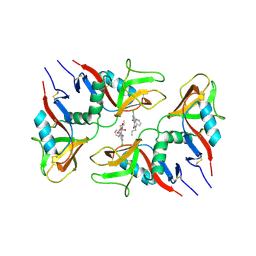 | | CRYSTAL STRUCTURE OF LECTIN-LIKE OX-LDL RECEPTOR 1 IN COMPLEX WITH BI-0115 | | Descriptor: | 9-chloranyl-5-propyl-11~{H}-pyrido[2,3-b][1,4]benzodiazepin-6-one, GLYCEROL, Oxidized low-density lipoprotein receptor 1 | | Authors: | Nar, H, Fiegen, D, Schnapp, G. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state
Commun Chem, 2020
|
|
1ZUX
 
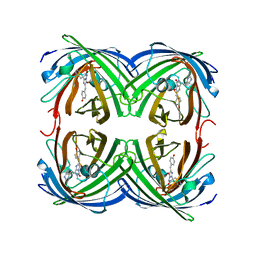 | | EosFP Fluorescent Protein- Green Form | | Descriptor: | green to red photoconvertible GPF-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AZU
 
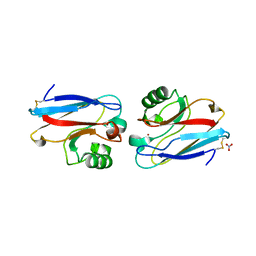 | |
2BTJ
 
 | | Fluorescent Protein EosFP - red form | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP | | Authors: | Nar, H, Nienhaus, K, Wiedenmann, J, Nienhaus, G.U. | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Photo-Induced Protein Cleavage and Green-to-Red Conversion of Fluorescent Protein Eosfp.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5AZU
 
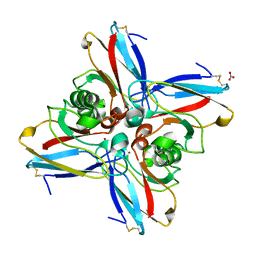 | |
6Y0F
 
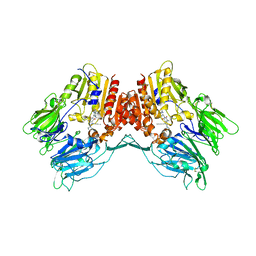 | | Structure of human FAPalpha in complex with linagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-[(3R)-3-Aminopiperidin-1-yl]-7-but-2-yn-1-yl-3-methyl-1-[(4-methylquinazolin-2-yl)methyl]-3,7-dihydro-1H-purine-2,6-d ione, ... | | Authors: | Nar, H, Schnapp, G, Schreiner, P. | | Deposit date: | 2020-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structure of human FAPalpha in complex with linagliptin
To Be Published
|
|
1GTQ
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE, ZINC ION | | Authors: | Nar, H, Huber, R, Heizmann, C.W, Thoeny, B, Buergisser, D. | | Deposit date: | 1995-09-16 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of 6-pyruvoyl tetrahydropterin synthase, an enzyme involved in tetrahydrobiopterin biosynthesis.
EMBO J., 13, 1994
|
|
4AZU
 
 | |
