3J89
 
 | | Structural Plasticity of Helical Nanotubes Based on Coiled-Coil Assemblies | | Descriptor: | synthetic peptide | | Authors: | Egelman, E.H, Xu, C, DiMaio, F, Magnotti, E, Modlin, C, Yu, X, Wright, E, Baker, D, Conticello, V.P. | | Deposit date: | 2014-10-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural plasticity of helical nanotubes based on coiled-coil assemblies.
Structure, 23, 2015
|
|
8GI2
 
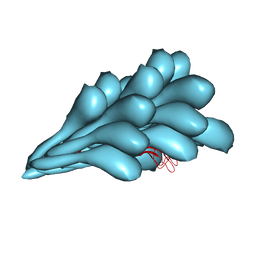 | | Cryo-EM structure of Natrinema sp. J7-2 Type IV pilus | | Descriptor: | Natrinema pilin, Orf10 | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Liu, Y, Krupovic, M, Egelman, E.H. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3LUE
 
 | | Model of alpha-actinin CH1 bound to F-actin | | Descriptor: | Actin, cytoplasmic 1, Alpha-actinin-3 | | Authors: | Galkin, V.E, Orlova, A, Salmazo, A, Djinovic-Carugo, K, Egelman, E.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Opening of tandem calponin homology domains regulates their affinity for F-actin.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6VY1
 
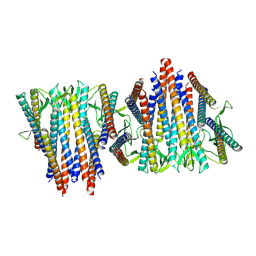 | | Cryo-EM structure of filamentous PFD from Methanocaldococcus jannaschii | | Descriptor: | Prefoldin subunit alpha 2 | | Authors: | Wang, F, Chen, Y.X, Ing, N.L, Hochbaum, A.I, Clark, D.S, Glover, D.J, Egelman, E.H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural Determination of a Filamentous Chaperone to Fabricate Electronically Conductive Metalloprotein Nanowires.
Acs Nano, 14, 2020
|
|
3BYH
 
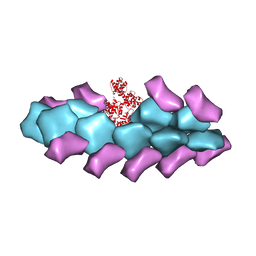 | | Model of actin-fimbrin ABD2 complex | | Descriptor: | Actin, fimbrin ABD2 | | Authors: | Galkin, V.E, Orlova, A, Cherepanova, O, Lebart, M.C, Egelman, E.H. | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6ANU
 
 | | Cryo-EM structure of F-actin complexed with the beta-III-spectrin actin-binding domain | | Descriptor: | Actin, cytoplasmic 1, Spectrin beta chain, ... | | Authors: | Wang, F, Orlova, A, Avery, A.W, Hays, T.S, Egelman, E.H. | | Deposit date: | 2017-08-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for high-affinity actin binding revealed by a beta-III-spectrin SCA5 missense mutation.
Nat Commun, 8, 2017
|
|
5O2Y
 
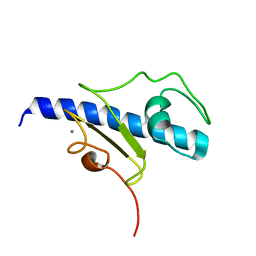 | | NMR structure of the calcium bound form of PulG, major pseudopilin from Klebsiella oxytoca T2SS | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Bardiaux, B, Vitorge, B, Thomassin, J.-L, Zheng, W, Yu, X, Egelman, E.H, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2017-05-23 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
5FLU
 
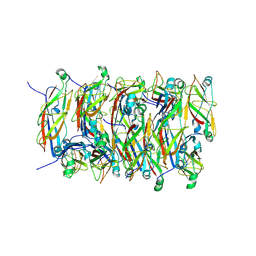 | | Structure of a Chaperone-Usher pilus reveals the molecular basis of rod uncoilin | | Descriptor: | PAP FIMBRIAL MAJOR PILIN PROTEIN | | Authors: | Hospenthal, M.K, Redzej, A, Dodson, K, Ukleja, M, Frenz, B, Hultgren, S.J, DiMaio, F, Egelman, E.H, Waksman, G. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Chaperone-Usher Pilus Reveals the Molecular Basis of Rod Uncoiling.
Cell(Cambridge,Mass.), 164, 2016
|
|
5A2T
 
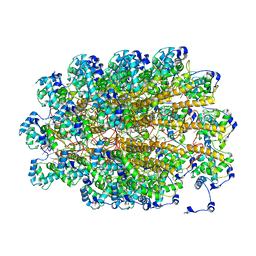 | | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses | | Descriptor: | BAMBOO MOSAIC VIRUS, COAT PROTEIN | | Authors: | DiMaio, F, Chen, C.C, Yu, X, Frenz, B, Hsu, Y.H, Lin, N.S, Egelman, E.H. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1N03
 
 | | Model for Active RecA Filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RecA protein | | Authors: | VanLoock, M.S, Yu, X, Yang, S, Lai, A.L, Low, C, Campbell, M.J, Egelman, E.H. | | Deposit date: | 2002-10-10 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | ATP-Mediated Conformational Changes in the RecA Filament
Structure, 11, 2003
|
|
6MKS
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | Chimera protein of NLR family CARD domain-containing protein 4 and EGFP | | Authors: | Zheng, W, Matyszewski, M, Sohn, J, Egelman, E.H. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the NLRC4CARDfilament provides insights into how symmetric and asymmetric supramolecular structures drive inflammasome assembly.
J. Biol. Chem., 293, 2018
|
|
7RX5
 
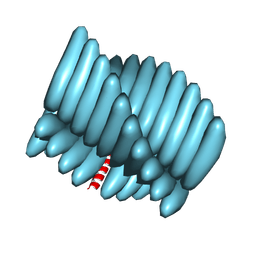 | | Cryo-EM reconstruction of Form1-N2 nanotube (Form I like) | | Descriptor: | F1-N2 nanotube | | Authors: | Wang, F, Gnewou, O.M, Solemanifar, A, Xu, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM of Helical Polymers.
Chem.Rev., 122, 2022
|
|
7RX4
 
 | | Cryo-EM reconstruction of AS2 nanotube (Form II like) | | Descriptor: | AS2 peptide | | Authors: | Wang, F, Gnewou, O.M, Solemanifar, A, Xu, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-08-21 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM of Helical Polymers.
Chem.Rev., 122, 2022
|
|
6HQE
 
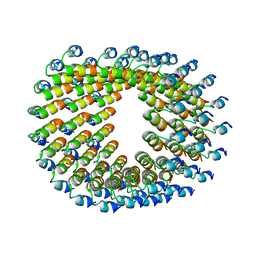 | | Cryo-EM of self-assembly peptide filament LRV_M3delta1 | | Descriptor: | peptide LRV_M3delta1 | | Authors: | Osinski, T, Wang, F, Hughes, S.A, Kreutzberger, M.A.B, Conticello, V.P, Egelman, E.H. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Ambidextrous helical nanotubes from self-assembly of designed helical hairpin motifs.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2QU4
 
 | | Model for Bacterial ParM Filament | | Descriptor: | Plasmid segregation protein parM | | Authors: | Orlova, A, Garner, E.C, Galkin, V.E, Heuser, J, Mullins, R.D, Egelman, E.H. | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | The structure of bacterial ParM filaments.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2REC
 
 | |
4BNE
 
 | | Pacsin2 Interacts with Membranes and Actin-Filaments | | Descriptor: | PROTEIN KINASE C AND CASEIN KINASE SUBSTRATE IN NEURONS PROTEIN 2, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Kostan, J, Salzer, U, Orlova, A, Toeroe, I, Hodnik, V, Schreiner, C, Merilainen, J, Nikki, M, Virtanen, I, Lehto, V.-P, Anderluh, G, Egelman, E.H, Djinovic-Carugo, K. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Direct Interaction of Actin Filaments with F-Bar Protein Pacsin2.
Embo Rep., 15, 2014
|
|
1Q5Z
 
 | | Crystal Structure of the C-terminal Actin Binding Domain of Salmonella Invasion Protein A (SipA) | | Descriptor: | SipA | | Authors: | Stebbins, C.E, Lilic, M, Galkin, V.E, Orlova, A, VanLoock, M.S, Egelman, E.H. | | Deposit date: | 2003-08-11 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms.
Science, 301, 2003
|
|
7JSV
 
 | | Cryo-EM structure of conjugative pili from carbapenem-resistant Klebsiella pneumoniae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, Pilin | | Authors: | Zheng, W, Pena, A, Frankel, G, Egelman, E.H. | | Deposit date: | 2020-08-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryoelectron-Microscopic Structure of the pKpQIL Conjugative Pili from Carbapenem-Resistant Klebsiella pneumoniae.
Structure, 28, 2020
|
|
7K3R
 
 | | Cryo-EM structure of AIM2-PYD filament | | Descriptor: | Interferon-inducible protein AIM2 | | Authors: | Zheng, W, Matyszewski, M, Sohn, J, Egelman, E.H. | | Deposit date: | 2020-09-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct axial and lateral interactions within homologous filaments dictate the signaling specificity and order of the AIM2-ASC inflammasome.
Nat Commun, 12, 2021
|
|
7KHW
 
 | |
8TJ2
 
 | | CryoEM structure of Myxococcus xanthus type IV pilus | | Descriptor: | Type IV major pilin protein PilA | | Authors: | Zheng, W, Egelman, E.H. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tight-packing of large pilin subunits provides distinct structural and mechanical properties for the Myxococcus xanthus type IVa pilus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6D5F
 
 | | Cryo-EM reconstruction of membrane-enveloped filamentous virus SFV1 (Sulfolobus filamentous virus 1) | | Descriptor: | DNA (336-MER), Fimbrial protein | | Authors: | Wang, F, Osinski, T, Liu, Y, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-08-29 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile.
Nat Commun, 9, 2018
|
|
6U7M
 
 | |
6C53
 
 | | Cryo-EM structure of the Type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Zheng, W, Wang, F, Luna-Rico, A, Francetic, O, Hultgren, S.J, Egelman, E.H. | | Deposit date: | 2018-01-13 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional role of the type 1 pilus rod structure in mediating host-pathogen interactions.
Elife, 7, 2018
|
|
