6NNT
 
 | |
6D0Q
 
 | | Structure of a DNA retention-prone PCNA variant | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Kelch, B.A, Gaubitz, C. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.80051017 Å) | | Cite: | Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex.
Nucleic Acids Res., 47, 2019
|
|
6WXE
 
 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Upright conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
8EM7
 
 | | Cryo-EM structure of LRP2 at pH 5.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
8EM4
 
 | | Cryo-EM structure of LRP2 at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Brasch, J, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
6XUX
 
 | | Crystal structure of Megabody Mb-Nb207-cYgjK_NO | | Descriptor: | CALCIUM ION, Nanobody,Glucosidase YgjK,Glucosidase YgjK,Nanobody | | Authors: | Steyaert, J, Uchanski, T, Fischer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.90000641 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
5J74
 
 | | Fluorogen activating protein AM2.2 in complex with TO1-2p | | Descriptor: | 1-(17-amino-5,8-dioxo-12,15-dioxa-4,9-diazaheptadecan-1-yl)-4-{[3-(3-sulfopropyl)-1,3-benzothiazol-3-ium-2-yl]methyl}quinolin-1-ium, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
6XV8
 
 | |
6MZN
 
 | |
6XTU
 
 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM | | Descriptor: | Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6XUT
 
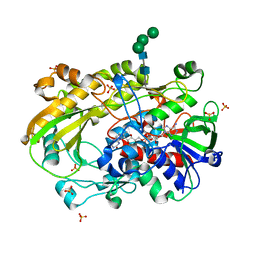 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Sciara, G, Vallone, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
8EQS
 
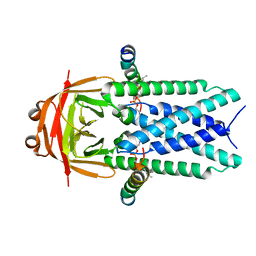 | | Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
4RWP
 
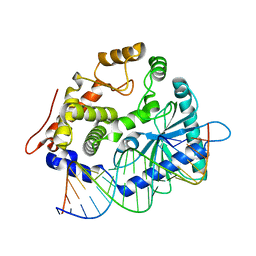 | | Crystal structure of porcine OAS1 in complex with dsRNA | | Descriptor: | 2'-5'-oligoadenylate synthase 1, RNA (5'-R(*GP*GP*CP*UP*UP*UP*UP*GP*AP*CP*CP*UP*UP*UP*AP*UP*GP*AP*A)-3'), RNA (5'-R(*UP*UP*CP*AP*UP*AP*AP*AP*GP*GP*UP*CP*AP*AP*AP*AP*GP*CP*C)-3') | | Authors: | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
6WZQ
 
 | |
6N87
 
 | | Plasmodium falciparum FVO apical membrane antigen 1 (AMA1) bound to MTSL spin-labelled cyclised RON2 peptide | | Descriptor: | Apical membrane antigen-1, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, backbone-cyclised peptide bcRON2hp | | Authors: | McGowan, S, Drinkwater, N. | | Deposit date: | 2018-11-28 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Identification of the Binding Site of Apical Membrane Antigen 1 (AMA1) Inhibitors Using a Paramagnetic Probe.
ChemMedChem, 14, 2019
|
|
6BYF
 
 | |
5KQA
 
 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | Descriptor: | GLUTATHIONE, Glutaredoxin-glutathione complex | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
6N60
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Microcin J25 (MccJ25) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-23 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3B9V
 
 | |
5KVC
 
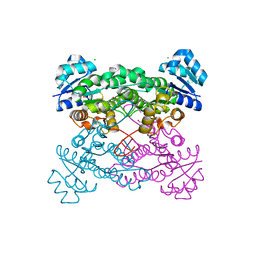 | |
6N9P
 
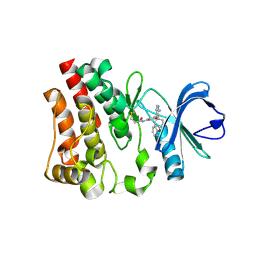 | |
6MWM
 
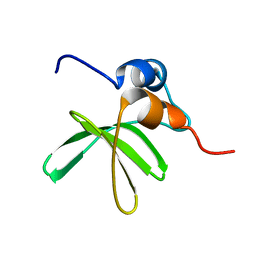 | | Bat coronavirus HKU4 SUD-C | | Descriptor: | Non-structural protein 3 | | Authors: | Staup, A.J, De Silva, I.U, Catt, J.T, Tan, X, Hammond, R.G, Johnson, M.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the SARS-Unique Domain C From the Bat Coronavirus HKU4.
Nat Prod Commun, 14, 2019
|
|
6WWZ
 
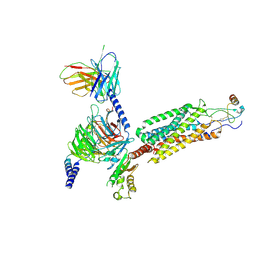 | | Cryo-EM structure of the human chemokine receptor CCR6 in complex with CCL20 and a Go protein | | Descriptor: | C-C chemokine receptor type 6,C-C chemokine receptor type 6, C-C motif chemokine 20, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wasilko, D.J, Johnson, Z.L, Ammirati, M, Han, S, Wu, H. | | Deposit date: | 2020-05-09 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for chemokine receptor CCR6 activation by the endogenous protein ligand CCL20.
Nat Commun, 11, 2020
|
|
5XTD
 
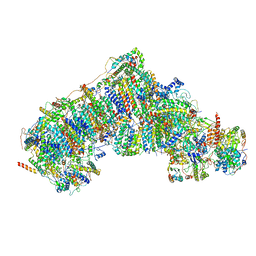 | | Cryo-EM structure of human respiratory complex I | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Acyl carrier protein, ... | | Authors: | Gu, J, Wu, M, Yang, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-08-30 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2.
Cell, 170, 2017
|
|
7QPJ
 
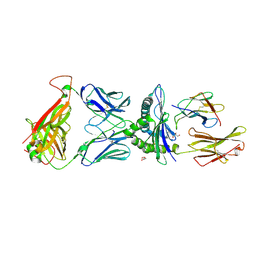 | | Crystal structure of engineered TCR (756) complexed to HLA-A*02:01 presenting MAGE-A10 9-mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, MHC class I antigen, ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
