2P98
 
 | | E. coli methionine aminopeptidase monometalated with inhibitor YE7 | | Descriptor: | IMIDAZO[2,1-A]ISOQUINOLINE-2-CARBOHYDRAZIDE, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
2P9A
 
 | | E. coli methionine aminopeptidase dimetalated with inhibitor YE6 | | Descriptor: | 5-(2-chlorophenyl)furan-2-carbohydrazide, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
2P99
 
 | | E. coli methionine aminopeptidase monometalated with inhibitor YE6 | | Descriptor: | 5-(2-chlorophenyl)furan-2-carbohydrazide, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q. | | Deposit date: | 2007-03-24 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Monometalated Methionine Aminopeptidase: Inhibitor Discovery and Crystallographic Analysis.
J.Med.Chem., 50, 2007
|
|
2Q96
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A18 | | Descriptor: | 5-(2-CHLOROBENZYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q94
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A04 | | Descriptor: | 5-[2-(TRIFLUOROMETHOXY)PHENYL]-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q93
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor B21 | | Descriptor: | 5-(2-METHOXYPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q95
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor A05 | | Descriptor: | 5-(2-CHLORO-4-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
2Q92
 
 | | E. coli methionine aminopeptidase Mn-form with inhibitor B23 | | Descriptor: | 5-(2-NITROPHENYL)-2-FUROIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z. | | Deposit date: | 2007-06-12 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of inhibition of E. coli methionine aminopeptidase: implication of loop flexibility in selective inhibition of bacterial enzymes.
Bmc Struct.Biol., 7, 2007
|
|
6WZO
 
 | |
6WZQ
 
 | |
6XI1
 
 | | Crystal structure of tetra-tandem repeat in extending RTX adhesin from Aeromonas hydrophila | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ye, Q, Vance, T.D.R, Conroy, B, Davies, P.L. | | Deposit date: | 2020-06-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Essential role of calcium in extending RTX adhesins to their target.
J Struct Biol X, 4, 2020
|
|
7N0I
 
 | |
6P8O
 
 | | Structure of P. aeruginosa ATCC27853 HORMA2-deltaC | | Descriptor: | CHLORIDE ION, HORMA domain containing protein, NICKEL (II) ION | | Authors: | Ye, Q, Corbett, K.D, Lau, R.K. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6U7B
 
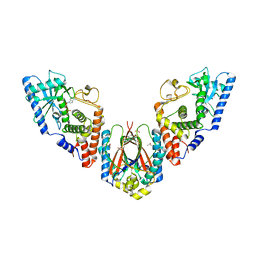 | | Structure of E. coli MS115-1 CdnC:HORMA-deltaN complex | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6UXF
 
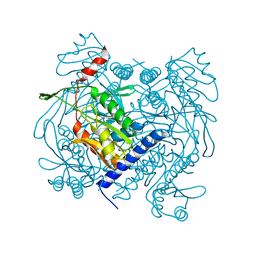 | | Structure of V. metoecus NucC, hexamer form | | Descriptor: | Vibrio meotecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6UXG
 
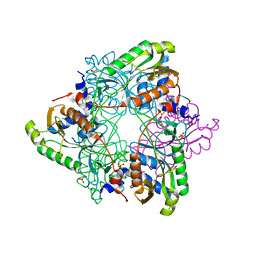 | | Structure of V. metoecus NucC, trimer form | | Descriptor: | SULFATE ION, Vibrio metoecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6XNR
 
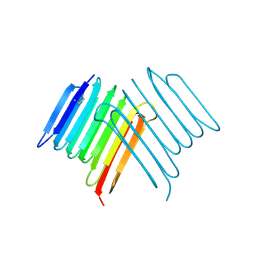 | | Crystal structure of Rhagium Mordax antifreeze protein | | Descriptor: | 1,2-ETHANEDIOL, Antifreeze protein | | Authors: | Ye, Q, Eves, R, Campbell, R.L, Davies, P.L. | | Deposit date: | 2020-07-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of an insect antifreeze protein reveals ordered waters on the ice-binding surface.
Biochem.J., 477, 2020
|
|
6XI3
 
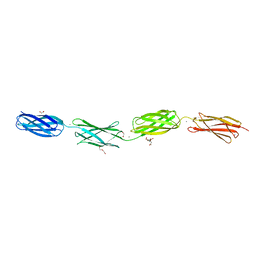 | | Crystal structure of tetra-tandem repeat in extending region of large adhesion protein | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ye, Q, Vance, T.D.R, Davies, P.L. | | Deposit date: | 2020-06-19 | | Release date: | 2020-10-14 | | Last modified: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Essential role of calcium in extending RTX adhesins to their target.
J Struct Biol X, 4, 2020
|
|
6P82
 
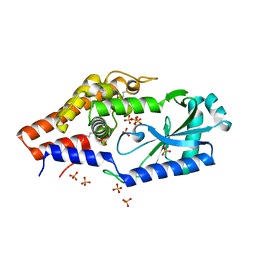 | | Structure of P. aeruginosa ATCC27853 CdnD, Apo form 1 | | Descriptor: | Nucleotidyltransferase, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8J
 
 | |
6P8S
 
 | |
7N0R
 
 | |
4JUQ
 
 | | Pseudomonas aeruginosa MetAP T2N mutant, in Mn form | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2013-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methionine excision captured by the structures of a methionine aminopeptidase
To be published
|
|
3LMI
 
 | |
4IL1
 
 | | Crystal Structure of the Rat Calcineurin | | Descriptor: | CALCIUM ION, Calmodulin, Calcineurin subunit B type 1, ... | | Authors: | Ye, Q, Faucher, F, Jia, Z. | | Deposit date: | 2012-12-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of calcineurin activation by calmodulin.
Cell Signal, 25, 2013
|
|
