3FAY
 
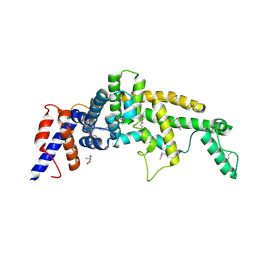 | | Crystal structure of the GAP-related domain of IQGAP1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ras GTPase-activating-like protein IQGAP1 | | Authors: | Kurella, V.B, Richard, J.M, Parke, C.L, Bellamy, H, Worthylake, D.K. | | Deposit date: | 2008-11-18 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the GTPase-activating protein-related domain from IQGAP1.
J.Biol.Chem., 284, 2009
|
|
1NF1
 
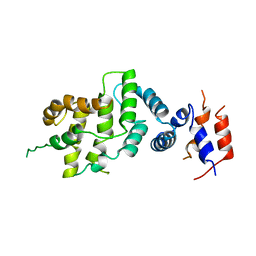 | | THE GAP RELATED DOMAIN OF NEUROFIBROMIN | | Descriptor: | PROTEIN (NEUROFIBROMIN) | | Authors: | Scheffzek, K, Ahmadian, M.R, Wiesmueller, L, Kabsch, W, Stege, P, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1998-07-08 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the GAP-related domain from neurofibromin and its implications.
EMBO J., 17, 1998
|
|
1WER
 
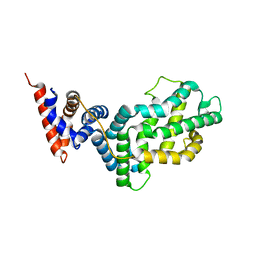 | | RAS-GTPASE-ACTIVATING DOMAIN OF HUMAN P120GAP | | Descriptor: | P120GAP | | Authors: | Scheffzek, K, Lautwein, A, Kabsch, W, Ahmadian, M.R, Wittinghofer, A. | | Deposit date: | 1996-11-20 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras.
Nature, 384, 1996
|
|
7R03
 
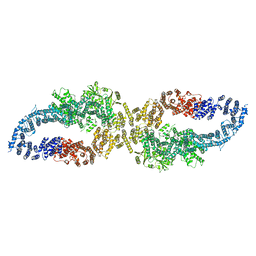 | |
7R04
 
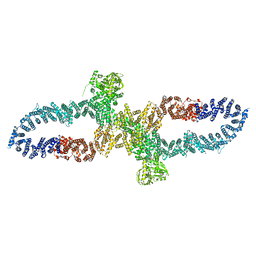 | | Neurofibromin in open conformation | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform I of Neurofibromin | | Authors: | Chaker-Margot, M, Scheffzek, K, Maier, T. | | Deposit date: | 2022-02-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of activation of the tumor suppressor protein neurofibromin.
Mol.Cell, 82, 2022
|
|
7MP6
 
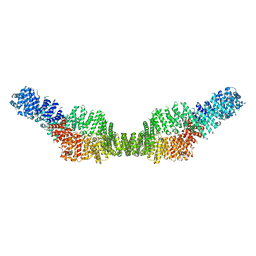 | | Neurofibromin homodimer | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MOC
 
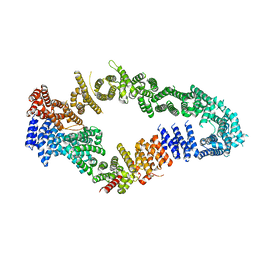 | | Neurofibromin core | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-01 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.56 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MP5
 
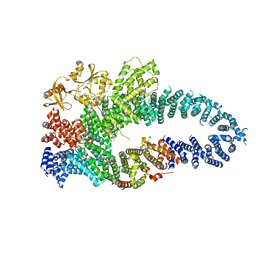 | | Autoinhibited neurofibrobmin | | Descriptor: | Isoform I of Neurofibromin | | Authors: | Lupton, C.J, Bayly-Jones, C, Ellisdon, A.M. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8E20
 
 | |
8EDO
 
 | |
8EDN
 
 | |
8EDL
 
 | |
8EDM
 
 | |
7PGU
 
 | | Autoinhibited structure of human neurofibromin isoform 2 stabilized by Zinc. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGT
 
 | | The structure of human neurofibromin isoform 2 in opened conformation. | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGS
 
 | | Consensus structure of human Neurofibromin isoform 2 | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGR
 
 | | The structure of human neurofibromin isoform 2 in closed conformation | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGP
 
 | |
7PGQ
 
 | | GAP-SecPH region of human neurofibromin isoform 2 in closed conformation. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
6OB2
 
 | | Crystal structure of wild-type KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | CHLORIDE ION, GLYCEROL, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3BXJ
 
 | | Crystal Structure of the C2-GAP Fragment of synGAP | | Descriptor: | Ras GTPase-activating protein SynGAP | | Authors: | Pena, V, Hothorn, M, Eberth, A, Kaschau, N, Parret, A, Gremer, L, Bonneau, F, Ahmadian, M.R, Scheffzek, K. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction.
Embo Rep., 9, 2008
|
|
5CJP
 
 | | The Structural Basis for Cdc42-Induced Dimerization of IQGAPs | | Descriptor: | Cell division cycle 42 (GTP binding protein, 25kDa), isoform CRA_a, ... | | Authors: | Worthylake, D.K, Boyapati, V.K, LeCour Jr, L. | | Deposit date: | 2015-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis for Cdc42-Induced Dimerization of IQGAPs.
Structure, 24, 2016
|
|
8BOS
 
 | |
9BZ4
 
 | |
6OB3
 
 | | Crystal structure of G13D-KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
