7ENA
 
 | | TFIID-based PIC-Mediator holo-complex in pre-assembled state (pre-hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENC
 
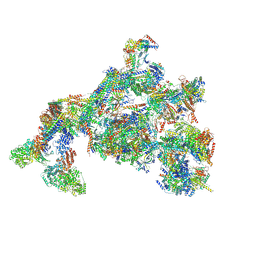 | | TFIID-based PIC-Mediator holo-complex in fully-assembled state (hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
5J44
 
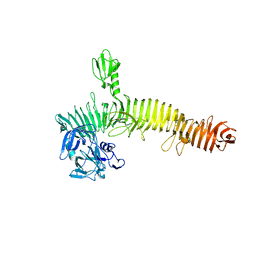 | | Crystal structure of the Secreted Extracellular protein A (SepA) from Shigella flexneri | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Serine protease SepA autotransporter | | Authors: | Birtley, J.R, Stern, L.J, McCormick, B, Maldonado-Contreras, A. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.912 Å) | | Cite: | Shigella depends on SepA to destabilize the intestinal epithelial integrity via cofilin activation.
Gut Microbes, 8, 2017
|
|
4MMZ
 
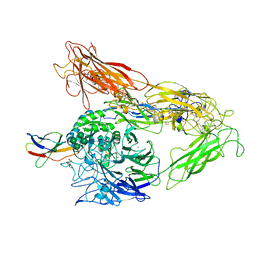 | | Integrin AlphaVBeta3 ectodomain bound to an antagonistic tenth domain of Fibronectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fibronectin, ... | | Authors: | van Agthoven, J, Xiong, J, Arnaout, M.A. | | Deposit date: | 2013-09-09 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for pure antagonism of integrin alpha V beta 3 by a high-affinity form of fibronectin.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8SCB
 
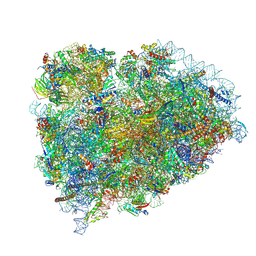 | | Terminating ribosome with SRI-41315 | | Descriptor: | (2S,4aS)-2-cyclobutyl-10-methyl-3-phenyl-2,10-dihydropyrimido[4,5-b]quinoline-4,5(3H,4aH)-dione, 18S_rRNA, 28S_rRNA, ... | | Authors: | Yip, M.C.J, Coelho, J.P.L, Oltion, K, Tauton, J, Shao, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center.
Nat.Chem.Biol., 20, 2024
|
|
8RJC
 
 | |
8RJD
 
 | |
8RJB
 
 | |
8QZZ
 
 | | Crystal structure of human eIF2 alpha-gamma complexed with PPP1R15A_420-452 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Substrate recruitment via eIF2 gamma enhances catalytic efficiency of a holophosphatase that terminates the integrated stress response.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8RKS
 
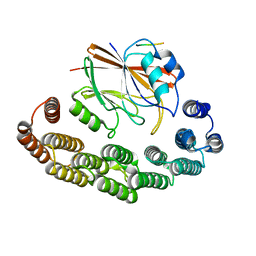 | | Structure of VPS29-VPS35 bound to the LFa motif R21 of Fam21. | | Descriptor: | Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35, WASH complex subunit 2A | | Authors: | Romano-Moreno, M, Astorga-Simon, E.N, Rojas, A.L, Hierro, A. | | Deposit date: | 2023-12-30 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Retromer-mediated recruitment of the WASH complex involves discrete interactions between VPS35, VPS29, and FAM21.
Protein Sci., 33, 2024
|
|
7UXH
 
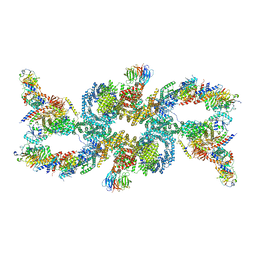 | | cryo-EM structure of the mTORC1-TFEB-Rag-Ragulator complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Cui, Z, Hurley, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the lysosomal mTORC1-TFEB-Rag-Ragulator megacomplex.
Nature, 614, 2023
|
|
7UXC
 
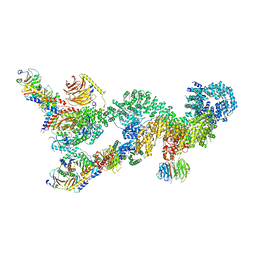 | |
4UAL
 
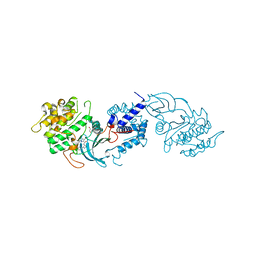 | | MRCK beta in complex with BDP00005290 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-1-(piperidin-4-yl)-N-[3-(pyridin-2-yl)-1H-pyrazol-4-yl]-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2014-08-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A novel small-molecule MRCK inhibitor blocks cancer cell invasion.
Cell Commun. Signal, 12, 2014
|
|
4UAK
 
 | | MRCK beta in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2014-08-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A novel small-molecule MRCK inhibitor blocks cancer cell invasion.
Cell Commun. Signal, 12, 2014
|
|
3REG
 
 | |
3REF
 
 | | Crystal structure of EhRho1 bound to GDP and Magnesium | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho-like small GTPase, ... | | Authors: | Bosch, D.E, Qiu, C, Siderovski, D.P. | | Deposit date: | 2011-04-04 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unique structural and nucleotide exchange features of the Rho1 GTPase of Entamoeba histolytica.
J.Biol.Chem., 286, 2011
|
|
7ROV
 
 | | KRAS G12D Mutant in complex with GMPPCP and cyclic peptide MP-9903 | | Descriptor: | Cyclic peptide MP-9903, GLYCEROL, Isoform 2B of GTPase KRas, ... | | Authors: | Orth, P. | | Deposit date: | 2021-08-02 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of cell active macrocyclic peptides with on-target inhibition of KRAS signaling.
Chem Sci, 12, 2021
|
|
7RSC
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSE
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
2PPZ
 
 | |
7R04
 
 | | Neurofibromin in open conformation | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform I of Neurofibromin | | Authors: | Chaker-Margot, M, Scheffzek, K, Maier, T. | | Deposit date: | 2022-02-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of activation of the tumor suppressor protein neurofibromin.
Mol.Cell, 82, 2022
|
|
7R03
 
 | |
2Q4G
 
 | | Ensemble refinement of the protein crystal structure of human ribonuclease inhibitor complexed with ribonuclease I | | Descriptor: | CITRIC ACID, Ribonuclease inhibitor, Ribonuclease pancreatic | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein.
J.Mol.Biol., 368, 2007
|
|
2PG1
 
 | |
4KED
 
 | |
