8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
8JT6
 
 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
8T7H
 
 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nanobody 43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-20 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T7C
 
 | | Crystal structure of human phospholipase C gamma 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2, CALCIUM ION | | Authors: | Chen, Y, Choi, H, Zhuang, N, Hu, L, Qian, D, Wang, J. | | Deposit date: | 2023-06-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal and cryo-EM structures of PLCg2 reveal dynamic inter-domain recognitions in autoinhibition
To Be Published
|
|
8PI2
 
 | |
8JSO
 
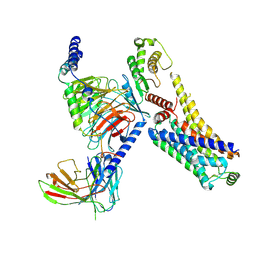 | | AMPH-bound hTAAR1-Gs protein complex | | Descriptor: | (2S)-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSP
 
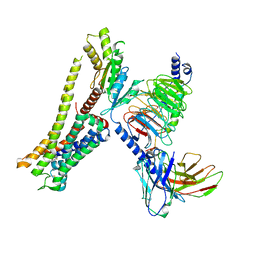 | | Ulotaront(SEP-363856)-bound Serotonin 1A (5-HT1A) receptor-Gi complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, 5-hydroxytryptamine receptor 1A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8T71
 
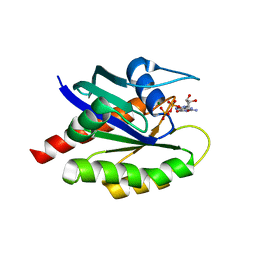 | | Crystal Structure of WT KRAS4a with bound GDP and Mg ion | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
8T72
 
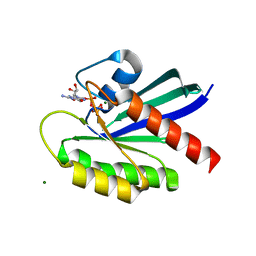 | | Crystal structure of WT KRAS4a with bound GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
8T75
 
 | |
8T73
 
 | | Crystal structure of KRAS4a-R151G with bound GDP and Mg ion | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Whitley, M.J, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-06-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparative analysis of KRAS4a and KRAS4b splice variants reveals distinctive structural and functional properties.
Sci Adv, 10, 2024
|
|
8T74
 
 | |
8PH7
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DPhF)(O2CCH3)3] in condition A | | Descriptor: | Lysozyme C, SODIUM ION, SUCCINIC ACID, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
8PH6
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K2[Ru2(DPhF)(CO3)3] in condition B | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C, Ru2(DPhF)(CO3)2(Formate), ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
8PH8
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DPhF)2(O2CCH3)2] in condition A | | Descriptor: | 1-oxidanyl-2,4,6,8-tetraphenyl-2,4,6,8-tetraza-1$l^{4},5$l^{3}-diruthenabicyclo[3.3.0]octane, Lysozyme C, SODIUM ION, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
8PH5
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DPhF)2(O2CCH3)2] in condition B | | Descriptor: | 1-oxidanyl-6,8,9,11-tetraphenyl-2,4-dioxa-6,8,9,11-tetraza-1$l^{5},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
8JS9
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Botulinum neurotoxin, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JS8
 
 | | Cryo-EM structure of SV2A in complex with BoNT/A2 Hc and levetiracetam | | Descriptor: | (2S)-2-(2-oxidanylidenepyrrolidin-1-yl)butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A. | | Deposit date: | 2023-06-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nat Commun, 15, 2024
|
|
8JRV
 
 | | Cryo-EM structure of the glucagon receptor bound to glucagon and beta-arrestin 1 | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), Glucagon, HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
8JRU
 
 | | Cryo-EM structure of the glucagon receptor bound to beta-arrestin 1 in ligand-free state | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, Nanobody 32, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
8JRL
 
 | | Crystal structure of P450 TleB with an indole alkaloid | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-(4-fluoranyl-1~{H}-indol-3-yl)-3-oxidanyl-propan-2-yl]-3-methyl-2-sulfanyl-butanamide, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, J, Yan, W.P. | | Deposit date: | 2023-06-17 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Regulation of P450 TleB catalytic flow for the synthesis of sulfur-containing indole alkaloids by substrate structure-directed strategy. and protein engineering.
Sci China Chem, 66, 2023
|
|
8T6J
 
 | | CDPPB-bound inactive mGlu5 | | Descriptor: | 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8PFV
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DAniF)(O2CCH3)3] in condition A | | Descriptor: | 9,11-bis(4-methoxyphenyl)-3,7-dimethyl-2,4,6,8-tetraoxa-9,11-diaza-1$l^{4},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, CHLORIDE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
8PFU
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K3[Ru2(CO3)4] in condition A | | Descriptor: | 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, CARBONATE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
8PFY
 
 | |
