5MP9
 
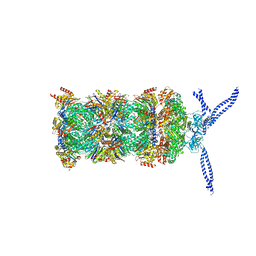 | | 26S proteasome in presence of ATP (s1) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8VLB
 
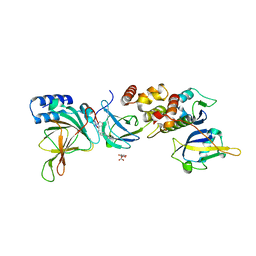 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 4 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A small molecule VHL molecular glue degrader for cysteine dioxygenase 1
To Be Published
|
|
8VL9
 
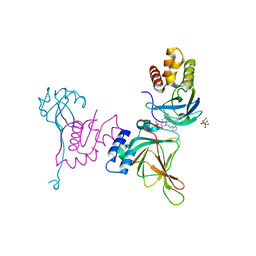 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 8 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A small molecule VHL molecular glue degrader for cysteine dioxygenase 1
To Be Published
|
|
8UHB
 
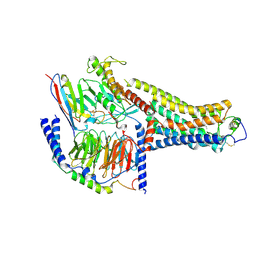 | | Cryo-EM Structure of the Ro5256390-bound hTA1-Gs heterotrimer signaling complex | | Descriptor: | (2R,4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zilberg, G, Warren, A.L, Parpounas, A.K, Wacker, D. | | Deposit date: | 2023-10-08 | | Release date: | 2024-01-10 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Molecular basis of human trace amine-associated receptor 1 activation.
Nat Commun, 15, 2024
|
|
8H1T
 
 | | Cryo-EM structure of BAP1-ASXL1 bound to chromatosome | | Descriptor: | DNA (187-MER), Histone H1.4, Histone H2A type 1-D, ... | | Authors: | Ge, W, Yu, C, Xu, R.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase.
Nature, 616, 2023
|
|
8H36
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H37
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.52 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7U50
 
 | |
6QDZ
 
 | | P38 alpha complex with AR117045 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{S})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P38 alpha complex with AR117045
To Be Published
|
|
6QE1
 
 | | P38 alpha complex with AR117046 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{R})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | P38 alpha complex with AR117045 and AR117046
To Be Published
|
|
8Q7R
 
 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | Descriptor: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
5E4V
 
 | | Crystal structure of measles N0-P complex | | Descriptor: | Nucleoprotein,Phosphoprotein | | Authors: | Guryanov, S.G, Liljeroos, L, Kasaragod, P, Kajander, T, Butcher, S.J. | | Deposit date: | 2015-10-07 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of the Measles Virus Nucleoprotein Core in Complex with an N-Terminal Region of Phosphoprotein.
J.Virol., 90, 2015
|
|
8JE2
 
 | | Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN | | Descriptor: | Cullin-2, Elongin-B, Elongin-C, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
6SFI
 
 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
5Y3R
 
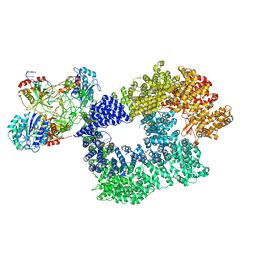 | | Cryo-EM structure of Human DNA-PK Holoenzyme | | Descriptor: | DNA (34-MER), DNA (36-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Yin, X, Liu, M, Tian, Y, Wang, J, Xu, Y. | | Deposit date: | 2017-07-29 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of human DNA-PK holoenzyme
Cell Res., 27, 2017
|
|
6SFJ
 
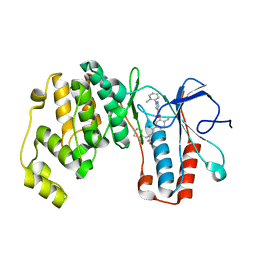 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
8W9C
 
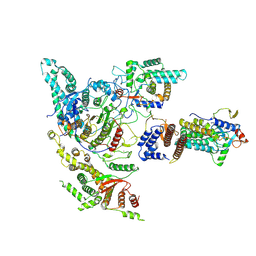 | |
3MEK
 
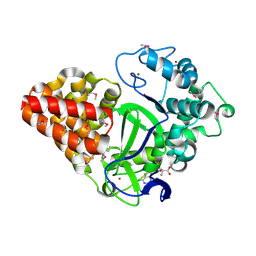 | | Crystal Structure of Human Histone-Lysine N-methyltransferase SMYD3 in Complex with S-adenosyl-L-methionine | | Descriptor: | S-ADENOSYLMETHIONINE, SET and MYND domain-containing protein 3, ZINC ION | | Authors: | Lam, R, Dombrovski, L, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Histone-Lysine N-methyltransferase SMYD3 in Complex with S-adenosyl-L-methionine
To be Published
|
|
6SFK
 
 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SFO
 
 | | MAPK14 with bound inhibitor SR-318 | | Descriptor: | 5-azanyl-~{N}-[[4-(3-cyclohexylpropylcarbamoyl)phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MAPK14 with bound inhibitor SR-318
To Be Published
|
|
8XVG
 
 | | Structure of human NuA4/TIP60 complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8IB8
 
 | | Human TRiC-PhLP2A-actin complex in the closed state | | Descriptor: | ACTB protein (Fragment), Phosducin-like protein 3, T-complex protein 1 subunit alpha, ... | | Authors: | Roh, S.H, Park, J, Kim, H, Lim, S. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A structural vista of phosducin-like PhLP2A-chaperonin TRiC cooperation during the ATP-driven folding cycle.
Nat Commun, 15, 2024
|
|
8R5H
 
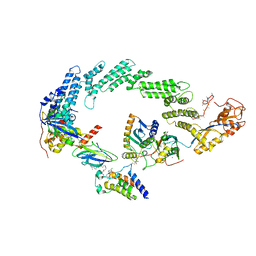 | | Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 5-azanylpentan-2-one, Bromodomain-containing protein 4, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-11-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
6TTU
 
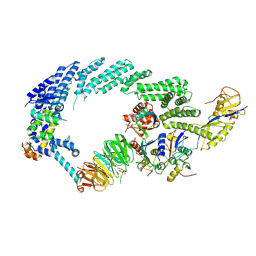 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | Descriptor: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
6O22
 
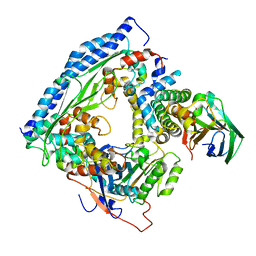 | | Structure of Asf1-H3:H4-Rtt109-Vps75 histone chaperone-lysine acetyltransferase complex with the histone substrate. | | Descriptor: | Histone H3.2, Histone H4, Histone acetyltransferase RTT109, ... | | Authors: | Danilenko, N, Carlomagno, T, Kirkpatrick, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun, 10, 2019
|
|
