8GNI
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
7NAK
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
3FY2
 
 | | Human EphA3 Kinase and Juxtamembrane Region Bound to Substrate KQWDNYEFIW | | Descriptor: | Ephrin type-A receptor 3, peptide substrate | | Authors: | Davis, T, Walker, J.R, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural recognition of an optimized substrate for the ephrin family of receptor tyrosine kinases.
Febs J., 276, 2009
|
|
3DZQ
 
 | | Human EphA3 kinase domain in complex with inhibitor AWL-II-38.3 | | Descriptor: | EPH receptor A3, N-[2-methyl-5-({[3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]carbonyl}amino)phenyl]isoxazole-5-carboxamide | | Authors: | Walker, J.R, Syeda, F, Gray, N, Mansoor, W, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinase Domain of Human Ephrin Type-A Receptor 3 (Epha3) in Complex with ALW-II-38-3.
To be Published
|
|
3FXX
 
 | | Human EphA3 Kinase and Juxtamembrane Region Bound to Substrate KQWDNYE[pTyr]IW | | Descriptor: | Ephrin type-A receptor 3, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Davis, T, Walker, J.R, Mackenzie, F, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural recognition of an optimized substrate for the ephrin family of receptor tyrosine kinases.
Febs J., 276, 2009
|
|
6ZG0
 
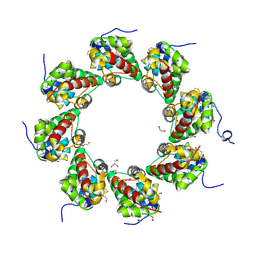 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
4P4C
 
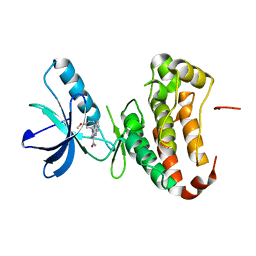 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(3-methoxyphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4G2F
 
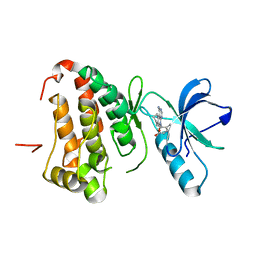 | | Human EphA3 kinase domain in complex with compound 7 | | Descriptor: | 1-amino-5-(5-hydroxy-2-methylphenyl)-7,8,9,10-tetrahydropyrimido[4,5-c]isoquinolin-6(5H)-one, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Discovery of a novel chemotype of tyrosine kinase inhibitors by fragment-based docking and molecular dynamics.
ACS MED.CHEM.LETT., 3, 2012
|
|
4GK3
 
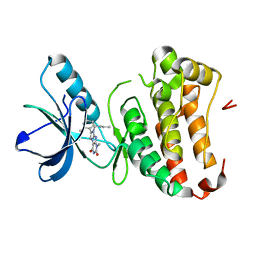 | | Human EphA3 Kinase domain in complex with ligand 87 | | Descriptor: | 8-butyl-1-methyl-7-(2-methylphenyl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
7CM5
 
 | | Full-length Sarm1 in a self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
4GK2
 
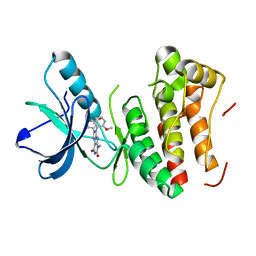 | | Human EphA3 Kinase domain in complex with ligand 66 | | Descriptor: | 7-(5-hydroxy-2-methylphenyl)-8-(2-methoxyphenyl)-1-methyl-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
7CM7
 
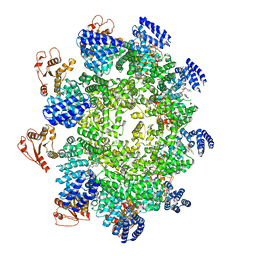 | | NAD+-bound Sarm1 E642A in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
4GK4
 
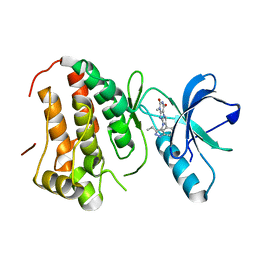 | | Human EphA3 Kinase domain in complex with ligand 90 | | Descriptor: | 8-butyl-1-methyl-7-(5-methyl-1H-indazol-4-yl)-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
7CM6
 
 | | NAD+-bound Sarm1 in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7DJT
 
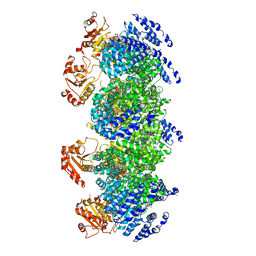 | | Human SARM1 inhibitory state bounded with inhibitor dHNN | | Descriptor: | NAD(+) hydrolase SARM1, O3-methyl O5-(2-methylpropyl) 2,6-dimethyl-4-[2-(oxidanylamino)phenyl]pyridine-3,5-dicarboxylate | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2020-11-21 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate.
Elife, 10, 2021
|
|
7ANW
 
 | | hSARM1 NAD+ complex | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Mim, C, Isupov, M.N, Zalk, R, Dessau, M, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-10-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
4P5Q
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(2-chlorophenyl)-N-(3-ethoxypropyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
4P5Z
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-[4-({[3-(trifluoromethyl)phenyl]carbamoyl}amino)phenyl]-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-20 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4TWO
 
 | | Human EphA3 Kinase domain in complex with compound 164 | | Descriptor: | 5-{[3-carbamoyl-4-(3,4-dimethylphenyl)-5-methylthiophen-2-yl]amino}-5-oxopentanoic acid, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-07-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases.
Acs Med.Chem.Lett., 6, 2015
|
|
2GSF
 
 | | The Human Epha3 Receptor Tyrosine Kinase and Juxtamembrane Region | | Descriptor: | Ephrin type-A receptor 3 | | Authors: | Walker, J.R, Davis, T, Dong, A, Newman, E.M, MacKenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure Of The Human Epha3 Receptor Tyrosine Kinase and Juxtamembrane Region
To be Published
|
|
2QOB
 
 | | Human EphA3 kinase domain, base structure | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2QOO
 
 | | Human EphA3 kinase and juxtamembrane region, Y596F:Y602F:Y742F triple mutant | | Descriptor: | Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
2QOF
 
 | | Human EphA3 kinase and juxtamembrane region, Y596F mutant | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
