6IC6
 
 | | Human cathepsin-C in complex with cyclopropyl peptidyl nitrile inhibitor 1 | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{R})-1-(aminomethyl)-2-[4-[4-(trifluoromethyl)phenyl]phenyl]cyclopropyl]-2-azanyl-butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
5EQS
 
 | | Crystal structure of a genotype 1a/3a chimeric HCV NS3/4A protease in complex with Asunaprevir | | Descriptor: | N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, NS3 protease, ZINC ION | | Authors: | Soumana, D, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Schiffer, C.A. | | Deposit date: | 2015-11-13 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Molecular and Dynamic Mechanism Underlying Drug Resistance in Genotype 3 Hepatitis C NS3/4A Protease.
J.Am.Chem.Soc., 138, 2016
|
|
6LUS
 
 | | Crystal structure of the Mengla Virus VP30 C-terminal domain | | Descriptor: | Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Wen, K.N, Chu, H.G, Wang, C.H, Qin, X.C. | | Deposit date: | 2020-01-30 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Mengla virus VP30 C-terminal domain.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6AO4
 
 | |
5LCW
 
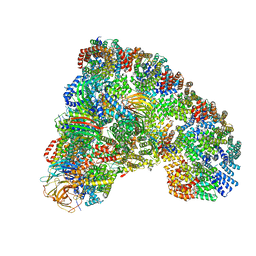 | | Cryo-EM structure of the Anaphase-promoting complex/Cyclosome, in complex with the Mitotic checkpoint complex (APC/C-MCC) at 4.2 angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Alfieri, C, Chang, L, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis of APC/C regulation by the spindle assembly checkpoint.
Nature, 536, 2016
|
|
6A7A
 
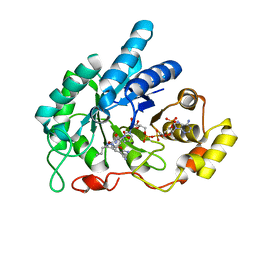 | | AKR1C1 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
5LMF
 
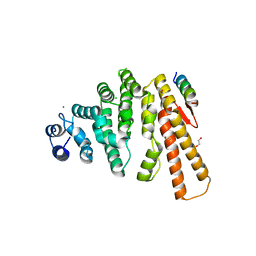 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM3 peptide (region 484-500) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6A7B
 
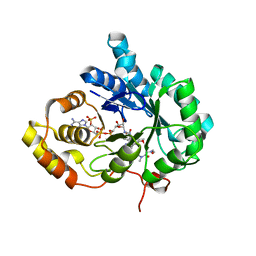 | | AKR1C3 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6S8Q
 
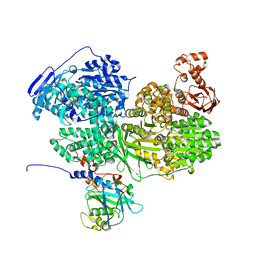 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, SULFATE ION, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Vester, K, Wahl, M.C. | | Deposit date: | 2019-07-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | The inactive C-terminal cassette of the dual-cassette RNA helicase BRR2 both stimulates and inhibits the activity of the N-terminal helicase unit.
J.Biol.Chem., 295, 2020
|
|
6S9I
 
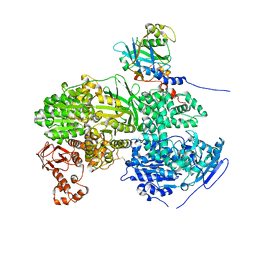 | |
4WHT
 
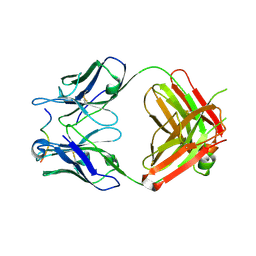 | |
6VIZ
 
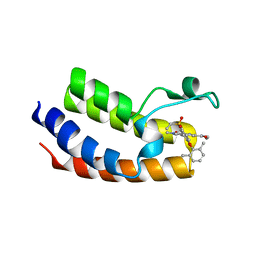 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
8FG0
 
 | |
5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
6VIY
 
 | | BRD2_Bromodomain2 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 2 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
8WQD
 
 | |
5LSJ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C delta-HEAD2 COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
6VIW
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
5ED7
 
 | | Crystal Structure of HSV-1 UL21 C-terminal Domain | | Descriptor: | CHLORIDE ION, Tegument protein UL21 | | Authors: | Metrick, C.M, Heldwein, E.E. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Novel Structure and Unexpected RNA-Binding Ability of the C-Terminal Domain of Herpes Simplex Virus 1 Tegument Protein UL21.
J.Virol., 90, 2016
|
|
5L9U
 
 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5EOC
 
 | | Crystal structure of Fab C2 in complex with a Cyclic variant of Hepatitis C Virus E2 epitope I | | Descriptor: | ALA-CYS-GLN-LEU-ILE-ASN-THR-ASN-GLY-SER-TRP-HIS-ILE-CYS, Fab fragment (Heavy chain), Fab fragment (Light chain) | | Authors: | Berisio, R, Ruggiero, A, Sandomenico, A, Patel, A.H, Ruvo, M, Vitagliano, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generation and Characterization of Monoclonal Antibodies against a Cyclic Variant of Hepatitis C Virus E2 Epitope 412-422.
J.Virol., 90, 2016
|
|
6XKC
 
 | | Crystal structure of E3 ligase | | Descriptor: | Protein fem-1 homolog C | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
3IOP
 
 | | PDK-1 in complex with the inhibitor Compound-8i | | Descriptor: | 2-(5-{[(2R)-2-amino-3-phenylpropyl]oxy}pyridin-3-yl)-8,9-dimethoxybenzo[c][2,7]naphthyridin-4-amine, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The identification of 8,9-dimethoxy-5-(2-aminoalkoxy-pyridin-3-yl)-benzo[c][2,7]naphthyridin-4-ylamines as potent inhibitors of 3-phosphoinositide-dependent kinase-1 (PDK-1).
Eur.J.Med.Chem., 45, 2010
|
|
7MWB
 
 | | ERAP1 binds peptide C-terminus of a SPF sequence (FKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,SPF Sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
7MWC
 
 | | ERAP1 binds peptide C-terminus of a LPF sequence (AAAAFKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,LPF sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
