9IJ2
 
 | | Cryo-EM Structure of MILI-piRNA-target (22-nt, comma) | | Descriptor: | Piwi-like protein 2, RNA (5'-R(P*CP*AP*AP*GP*UP*UP*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*A)-3'), RNA (5'-R(P*UP*UP*AP*CP*CP*AP*UP*CP*AP*AP*CP*AP*UP*GP*GP*AP*AP*AP*CP*UP*UP*G)-3') | | Authors: | Li, Z.Q, Xu, Q.K, Wu, J.P, Shen, E.Z. | | Deposit date: | 2024-06-21 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into RNA cleavage by PIWI Argonaute.
Nature, 639, 2025
|
|
5TGM
 
 | | Crystal structure of the S.cerevisiae 80S ribosome in complex with the A-site bound aminoacyl-tRNA analog ACCA-Pro | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Melnikov, S, Mailliot, J, Yusupov, M. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular insights into protein synthesis with proline residues.
EMBO Rep., 17, 2016
|
|
1ZHO
 
 | | The structure of a ribosomal protein L1 in complex with mRNA | | Descriptor: | 50S ribosomal protein L1, POTASSIUM ION, mRNA | | Authors: | Nevskaya, N, Tishchenko, S, Volchkov, S, Kljashtorny, V, Nikonova, E, Nikonov, O, Nikulin, A, Kohrer, C, Piendl, W, Zimmermann, R, Stockley, P, Garber, M, Nikonov, S. | | Deposit date: | 2005-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights into the interaction of ribosomal protein L1 with RNA.
J.Mol.Biol., 355, 2006
|
|
5K77
 
 | | Dbr1 in complex with 7-mer branched RNA | | Descriptor: | FE (II) ION, HYDROXIDE ION, RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Metal dependence and branched RNA cocrystal structures of the RNA lariat debranching enzyme Dbr1.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4UYK
 
 | | Crystal structure of a Signal Recognition Particle Alu domain in the elongation arrest conformation | | Descriptor: | SIGNAL RECOGNITION PARTICLE 14 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, SRP RNA | | Authors: | Bousset, L, Mary, C, Brooks, M.A, Scherrer, A, Strub, K, Cusack, S. | | Deposit date: | 2014-09-01 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation.
RNA, 20, 2014
|
|
7V6C
 
 | | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | | Descriptor: | Dicer-2, isoform A, R2D2, ... | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
5HW4
 
 | | Crystal structure of Escherichia coli 16S rRNA methyltransferase RsmI in complex with AdoMet | | Descriptor: | Ribosomal RNA small subunit methyltransferase I, S-ADENOSYLMETHIONINE | | Authors: | Zhao, M, Zhang, H, Dong, Y, Gong, Y. | | Deposit date: | 2016-01-28 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Structural Insights into the Methylation of C1402 in 16S rRNA by Methyltransferase RsmI
Plos One, 11, 2016
|
|
6HAK
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with a double stranded RNA represents the RT transcription initiation complex prior to nucleotide incorporation | | Descriptor: | Gag-Pol polyprotein, MAGNESIUM ION, RNA (5'-R(P*AP*GP*UP*GP*GP*CP*GP*GP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2018-08-07 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of HIV-1 RT/dsRNA initiation complex prior to nucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2A2E
 
 | | Crystal structure of the RNA subunit of Ribonuclease P. Bacterial A-type. | | Descriptor: | OSMIUM ION, RNA subunit of RNase P | | Authors: | Torres-Larios, A, Swinger, K.K, Krasilnikov, A.S, Pan, T, Mondragon, A. | | Deposit date: | 2005-06-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the RNA component of bacterial ribonuclease P.
Nature, 437, 2005
|
|
3J79
 
 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, large subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
2DFX
 
 | | Crystal structure of the carboxy terminal domain of colicin E5 complexed with its inhibitor | | Descriptor: | Colicin-E5, Colicin-E5 immunity protein | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-03-06 | | Release date: | 2007-01-23 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
2DJH
 
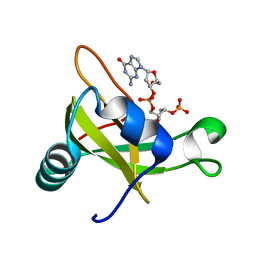 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-04-03 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
5DET
 
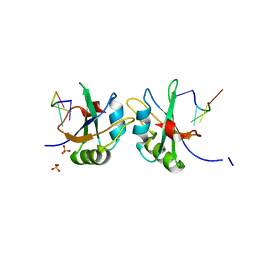 | | X-ray structure of human RBPMS in complex with the RNA | | Descriptor: | RNA (5'-R(*UP*CP*AP*C)-3'), RNA (5'-R(P*UP*CP*AP*CP*U)-3'), RNA-binding protein with multiple splicing, ... | | Authors: | Teplova, M, Farazi, T.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
9FSS
 
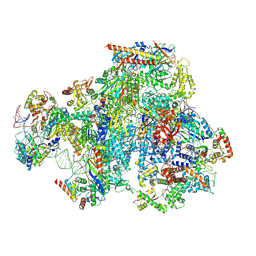 | | RNA Polymerase III Class III Open Mini Pre-Initiation complex 2 (OC2-mini) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
9FSP
 
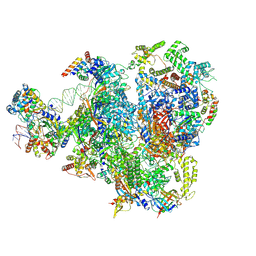 | | RNA Polymerase III Class III Open Pre-initation Complex 2 (OC2) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
9FSR
 
 | | RNA Polymerase III Class III Open Mini Pre-initiation Complex 1 (OC1-mini) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
9FSQ
 
 | | RNA Polymerase III Class III Melting Pre-Initiation Complex (MC) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Shah, S.Z, Ramsay, E.P, Cecatiello, V, Perry, T.N, Vannini, A. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into distinct mechanisms of RNA polymerase II and III recruitment to snRNA promoters.
Nat Commun, 16, 2025
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
5D0A
 
 | | Crystal structure of epoxyqueuosine reductase with cleaved RNA stem loop | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, GLYCEROL, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Bandarian, V, Drennan, C.L. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
7DD3
 
 | | Cryo-EM structure of the pre-mRNA-loaded DEAH-box ATPase/helicase Prp2 in complex with Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2, pre-mRNA | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
8OEF
 
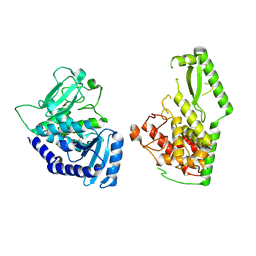 | |
5ZYA
 
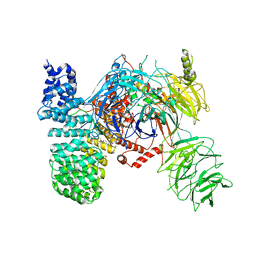 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
1OB5
 
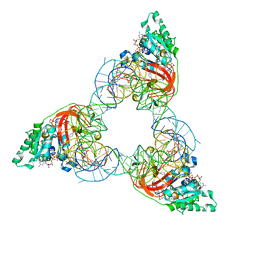 | | T. aquaticus elongation factor EF-Tu complexed with the antibiotic enacyloxin IIa, a GTP analog, and Phe-tRNA | | Descriptor: | ELONGATION FACTOR TU, ENACYLOXIN IIA, MAGNESIUM ION, ... | | Authors: | Dahlberg, C, Nielsen, R.C, Parmeggiani, A, Nyborg, J, Nissen, P. | | Deposit date: | 2003-01-24 | | Release date: | 2005-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Enacyloxin Iia Pinpoints a Binding Pocket of Elongation Factor TU for Development of Novel Antibiotics.
J.Biol.Chem., 281, 2006
|
|
1XMO
 
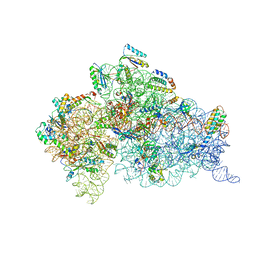 | | Crystal Structure of mnm5U34t6A37-tRNALysUUU Complexed with AAG-mRNA in the Decoding Center | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Ramakrishnan, V, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The role of modifications in codon discrimination by tRNA(Lys)(UUU).
Nat.Struct.Mol.Biol., 11, 2004
|
|
